Improving Microbiome Data Reproducibility Across Labs
The surge of microbiome research in the past decade has generated a wealth of data to be explored. But, a recent editorial in Nature Microbiology1 puts a spotlight on the obstacles that impede microbiome data sharing and comparison.
One major obstacle stems from the variations caused by the use of different methods and tools. The author writes, “The range of extraction methods, technologies and analytical pipelines available for processing similar samples makes comparison between studies incredibly difficult.”
An NGS-based microbiome workflow consists of multiple steps including sample collection, DNA extraction, library preparation, NGS sequencing, and bioinformatics analysis with each method or research tool varying significantly in performance which potentially introduces substantial bias and error.
Two studies have reported similar issues: the Microbiome Quality Control Project (MBQC) 3 found “studies have been difficult to reproduce across investigations” and a publication4 in Nature Microbiology assessing US microbiome research reported that “variation at each step of the pipeline is enormous”.
As a result, direct comparison between labs is nearly impossible. A prominent example of this issue is the discrepant data between American Gut’s and µBiome’s analyses of the same fecal sample2 (Figure 1).
As pointed out by the recent editorial in Nature Microbiology 1, global standardization of microbiome workflows is a distant aim, given constant technological advances. However, the best way to improve data reproducibility is to continually improve the accuracy of microbiome measurements across the field through the use of benchmarking materials (e. g. microbiome reference materials) and better research tools (e. g. unbiased DNA extraction methods). With continual improvements, bias and error will be reduced to the point that comparison will be realistic even if there are subtle differences, versus the present, where the most cited extraction method has been shown to induce misrepresentation of organisms by orders of magnitude. To learn more about the challenges facing the field of Microbiomics click here to download our Newsletter.
Also, click here to learn how our ZymoBIOMICS™ Microbial Community Standard, ZymoBIOMICS™ DNA Miniprep, and other microbiomics products can help you improve the accuracy of your microbiome measurements.
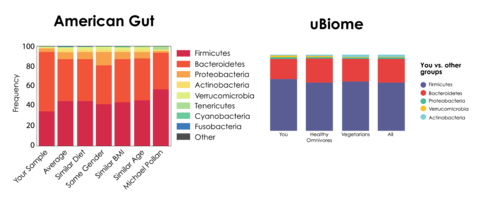
References:
1 Overcoming hurdles in sharing microbiome data. Nature microbiology 2, 1573-1573, doi:10.1038/s41564-017-0077-3 (2017).
2 Saey, T. H. Here's the poop on getting your gut microbiome analyzed, (2014).
3 Sinha, R., Abnet, C. C., White, O., Knight, R. & Huttenhower, C. The microbiome quality control project: baseline study design and future directions. Genome biology 16, 276, doi:10.1186/s13059-015-0841-8 (2015).
4 Stulberg, E. et al. An assessment of US microbiome research. Nature microbiology 1, 15015, doi:10.1038/nmicrobiol.2015.15 (2016).