Shotgun metagenomic sequencing has widely been used to characterize an assortment of microbiome samples,
including those derived from human/animal body sites and environmental samples (e.g. soil, sediments, sludge, water, air, etc.).
The use of shotgun sequencing for metagenomics, rather than a targeted microbiome profiling (e.g. 16S/ITS),
enables more information to be obtained from a sample. This includes cross-kingdom microbial identification, metabolic
function profiling, strain-level differentiation, detection of virulence genes and antibiotic resistance genes.
The workflow of shotgun metagenomic sequencing is complex with multiple steps where improper handling can cause
bias that can significantly alter the results. A well-established workflow with extensive validation is crucial to
the success of a metagenomic sequencing project.
Zymo Research is a well-known pioneer as a reagent and kit provider for microbiome research. Our innovative
microbiome products include DNA/RNA Shield for unbiased sample collection, ZymoBIOMICS DNA kits for unbiased lysis, and
microbiome standards to ensure superior process quality control.
Building on the top of these innovative products, we have established a shotgun metagenomic sequencing service
that was optimized thoroughly and validated to ensure unbiased microbiome profiling.
Our shotgun metagenomic sequencing service also includes a comprehensive bioinformatics report that includes
strain-level taxonomy profiling, metabolic gene family and metabolic pathway profiling, virulence gene and
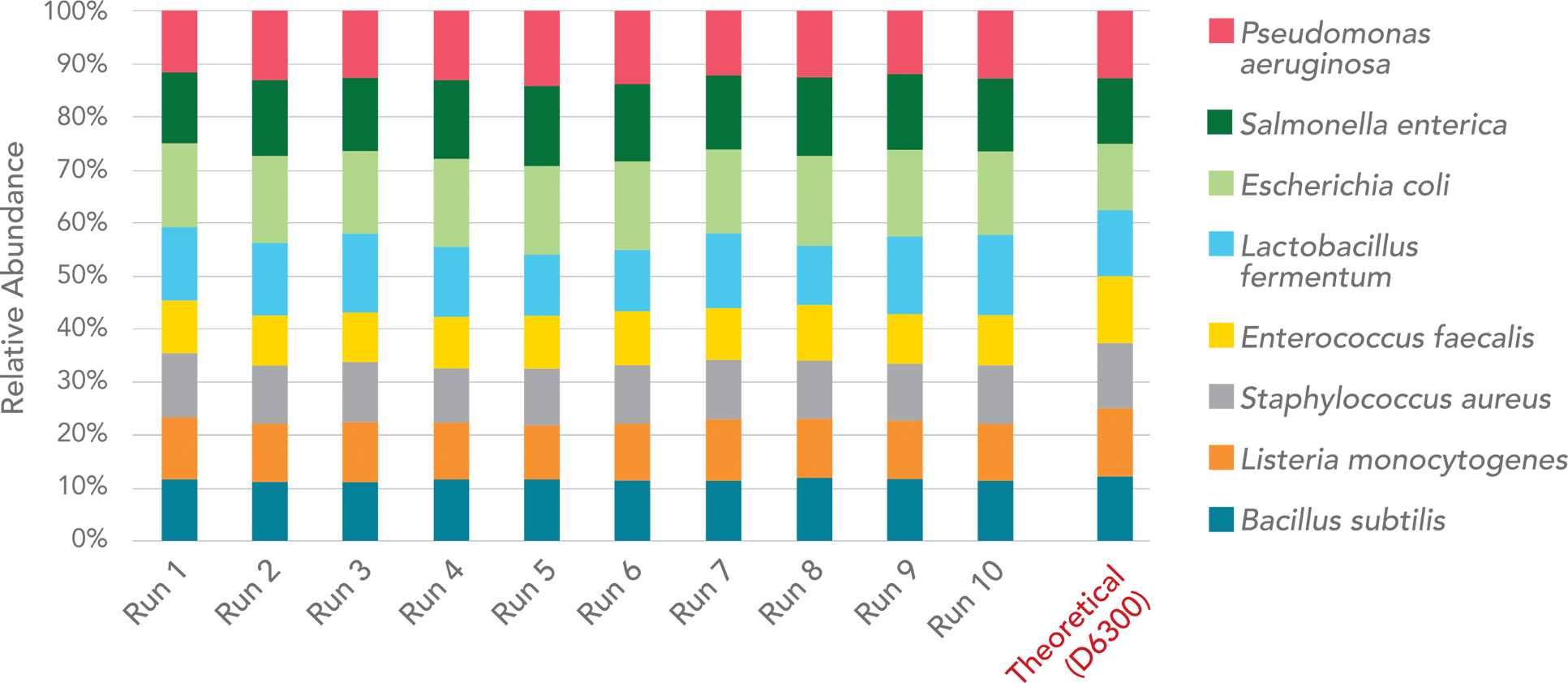
antibiotics resistance gene identification, statistical analysis, etc. Innovations on the taxonomy assignment algorithm
enable us to dramatically reduce the false positives in microbial identification, a common issue that most existing
metagenomics tools and most service providers suffer from.
Academic researchers and industrial clients alike trust our shotgun metagenomic sequencing service. This
includes high-throughput direct-to-consumer testing companies that also enjoy our fast turn-around time and the complete
workflow from sample collection to bioinformatic analysis.