Successfully Added to Cart
Updated Cart Total:
0
items
Subtotal:
$0.00 USD
Customers also bought...
-
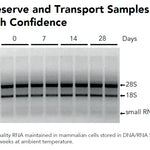 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
- Two minute DNA clean-up from any step in a standard ChIP protocol.
- DNA is ideal for PCR, arrays, DNA quantification, Southern blot analysis, sequencing, and other molecular applications.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Two minute DNA clean-up from any step in a standard ChIP protocol.
- DNA is ideal for PCR, arrays, DNA quantification, Southern blot analysis, sequencing, and other molecular applications.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | Price | |
|---|---|---|---|---|
| D3004-4-10 | DNA Elution Buffer | 10 ml | $17.00 | |
| D4003-2-6 | DNA Wash Buffer (Concentrate) | 6 ml | $12.00 | |
| C1004-50 | Zymo-Spin IC Columns | 50 Pack | $64.00 | |
| C1003-50 | Zymo-Spin I Columns | 50 Pack | $60.00 | |
| C1001-50 | Collection Tubes | 50 Pack | $18.00 | |
| D5201-1-50 | ChIP DNA Binding Buffer | 50 ml | $40.00 | |
Description
Performance
Technical Specifications
| Elution Volume | ≥ 6 µl |
|---|---|
| Processing Time | 2 minutes |
| Recovery | 70-90% for DNA 50 bp - 10 kb 70% for DNA>10 kb |
| Size Range | 50 bp - 23 kb |
| Storage | All components should be stored at room temperature. |
Resources
Documents
Citations
Need help? Contact Us




