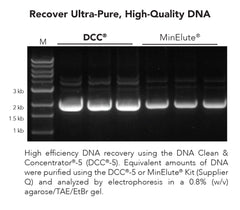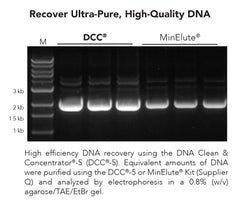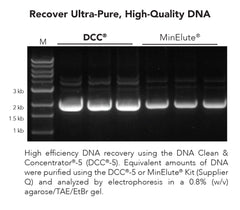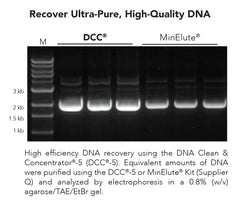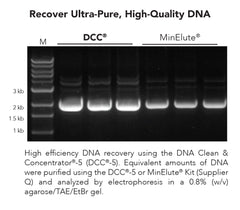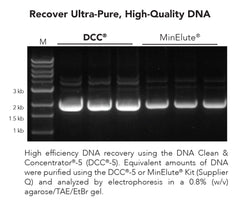
DNA Clean & Concentrator-5
The DNA Clean & Concentrator-5 is a PCR purification kit that provides purification of up to 5 µg DNA from PCR, endonuclease digestions, DNA modification...
D4004, D4003, D4014, D4013, D4003T
Comparison Table
| Catalog # | Product | Binding Capacity | Elution Volume | Format |
|---|---|---|---|---|
| D4013 | DNA Clean & Concentrator-5 | 5 µg | ≥ 6 µl | Spin-Column |
| D4033 | DNA Clean & Concentrator-25 | 25 µg | ≥ 25 µl | Spin-Column |
| D4029 | DNA Clean & Concentrator-100 | 100 µg | ≥ 150 µl | Spin-Column |
| D4031 | DNA Clean & Concentrator-500 | 500 µg | ≥ 2 ml | Spin-Column |
| D4012 | DNA Clean & Concentrator MagBead Kit | 4 µg | ≥ 30 µl | Magnetic Bead |
| D4017 | ZR-96 DNA Clean-up Kit | 5 µg | ≥ 30 µl | 96-Well Plate |
| D4023 | ZR-96 DNA Clean & Concentrator-5 (Deep Well) | 5 µg | ≥ 10 µl | 96-Well Plate |
Uses for PCR Purification Kits
Use PCR purification kits to remove enzymes, nucleotides, primers, and salts. These are present in PCR and other enzymatic reactions before using the DNA downstream. PCR and other enzymatic reactions serve as intermediate steps in many common molecular biology techniques. Sequencing, cloning, and DNA modification are examples of these. It is critical that the DNA is high-quality and free of any potential inhibitors for downstream use.
We believe to have developed the most comprehensive technologies for DNA clean-up and concentration from any preparation. The total removal of salts/alcohol from samples with uniquely designed spin-columns and plates is core to these products. Specifically, this ensures complete elution with no binding/wash buffer carryover. Coupled with uniquely formulated buffers, our PCR purification kits assure the purification of high-quality DNA without the inclusion of inhibitors.
Tips to Find the Best Kits
First, many molecular biology techniques are sensitive to certain salts, proteins and organic compounds. Therefore, it is imperative that PCR purification kits produce DNA that is ultra-pure and free of inhibitors that can interfere downstream.
Second, the volume of DNA that can be used in downstream reactions is limited, thus the DNA recovered from the PCR Purification kit needs to be highly concentrated.
Third, downstream techniques also require a minimum amount of input DNA. The kit should not lose a significant amount of DNA during the purification process.
Fourth, the protocol needs to be fast and easy to perform. Perform PCR purification on many samples simultaneously.
Finally, many different types of DNA reactions often need to be purified in molecular biology workflows, hence you want a PCR purification kit that can clean-up a wide range of sample types.
Do I Need a PCR Purification Kit?
Traditionally, DNA was purified from PCR and other enzymatic reactions using phenol-chloroform extraction and alcohol precipitation. However, this method required the use of hazardous organic reagents that need special care when handling and disposing. In addition, the phase separation and alcohol precipitation steps are long and tedious to perform and prone to errors. Typically, a significant amount of DNA is lost during the extraction process. The resulting DNA often contains inhibitors.
We've pioneered rapid, efficient PCR purification kits with the introduction of our DNA Clean & Concentrator (DCC) product line. These PCR purification kits utilize a uniquely formulated DNA binding chemistry and novel spin-column design. This formulation rapidly purifies ultra-pure DNA from PCR and other enzymatic reactions without the use of hazardous chemicals. Since its inception, the DCC family of PCR purification kits have evolved into one of the most efficient and versatile methods for cleaning and concentrating DNA. Most importantly, from a range of sample sources into minimal elution volumes (i.e., ≥ 6 µl).
Use DCC Kits or Gel Extraction Kits?
Both PCR purification kits and Gel extraction kits remove enzymes, nucleotides, primers, and salts present in PCR and other enzymatic reactions. However, PCR purification kits will be much faster and easier to perform. There is no requirement to resolve the DNA product using gel electrophoresis. However, if unwanted DNA products are also produced during the reaction, a Gel extraction kit must be employed to selectively purify the DNA fragment of interest.
Uses for PCR Purification Kits
Use PCR purification kits to remove enzymes, nucleotides, primers, and salts. These are are present in PCR and other enzymatic reactions before using the DNA downstream. PCR and other enzymatic reactions serve as intermediate steps in many common molecular biology techniques. Sequencing, cloning and DNA modification are examples of these. It is critical that the DNA is high-quality and free of any potential inhibitors for downstream use.
We believe to have developed the most comprehensive technologies for DNA clean-up and concentration from any preparation. The total removal of salts/alcohol from samples with uniquely designed spin-columns and plates is core to these products. Specifically, this ensures complete elution with no binding/wash buffer carryover. Coupled with uniquely formulated buffers, our PCR purification kits assure the purification of high-quality DNA without the inclusions of inhibitors.
Tips to Find the Best Kits
First, many molecular biology techniques are sensitive to certain salts, proteins and organic compounds. Therefore, it is imperative that PCR purification kits produce DNA that is ultra-pure and free of inhibitors that can interfere downstream.
Second, the volume of DNA that can be used in downstream reactions is limited, thus the DNA recovered from the PCR Purification kit needs to be highly concentrated.
Third, downstream techniques also require a minimum amount of input DNA. The kit should not lose a significant amount of DNA during the purification process.
Fourth, the protocol needs to be fast and easy to perform. Perform PCR purification on many samples simultaneously.
Finally, many different types of DNA reactions often need to be purified in molecular biology workflows, hence you want a PCR purification kit that can clean-up a wide range of sample types.
Do I Need a PCR Purification Kit?
Traditionally, DNA was purified from PCR and other enzymatic reactions using phenol-chloroform extraction and alcohol precipitation. However, this method required the use of hazardous organic reagents that need special care when handling and disposing. In addition, the phase separation and alcohol precipitation steps are long and tedious to perform and prone to errors. Typically, a significant amount of DNA is lost during the extraction process. The resulting DNA often contains inhibitors.
We've pioneered rapid, efficient PCR purification kits with the introduction of our DNA Clean & Concentrator (DCC) product line. These PCR purification kits utilize a uniquely formulated DNA binding chemistry and novel spin-column design. This formulation rapidly purifies ultra-pure DNA from PCR and other enzymatic reactions without the use of hazardous chemicals. Since its inception, the DCC family of PCR purification kits have evolved into one of the most efficient and versatile methods for cleaning and concentrating DNA. Most importantly, from a range of sample sources into minimal elution volumes (i.e., ≥ 6 µl).
Use DCC Kits or Gel Extraction Kits?
Both PCR purification kits and Gel extraction kits remove enzymes, nucleotides, primers, and salts present in PCR and other enzymatic reactions. However, PCR purification kits will be much faster and easier to perform. There is no requirement to resolve the DNA product using gel electrophoresis. However, if unwanted DNA products are also produced during the reaction, a Gel extraction kits must be employed to selectively purify the DNA fragment of interest.