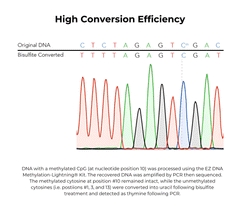
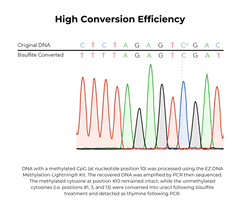
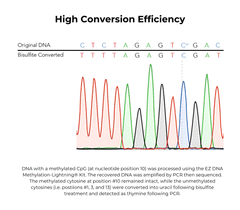
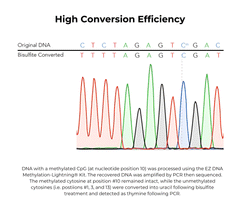
EZ DNA Methylation-Lightning Kits
The EZ DNA Methylation-Lightning Kits are citation-leading bisulfite conversion kits that allow the fastest complete bisulfite conversion of DNA, featuring rapid and reliable bisulfite treatment for methylation analysis. A key feature of these bisulfite conversion kits is the ready-to-use Lightning Conversion Reagent. No preparation is necessary, simply add this unique reagent to a DNA sample, incubate for approximately 1 hour, and let the reaction proceed to completion. These bisulfite conversion kits allow desulphonation and clean-up of converted DNA to be performed using a unique low-elution spin column. In addition to the spin-column format, the EZ-96 DNA Methylation-Lightning MagPrep Kit is the fastest magnetic bead-based bisulfite conversion kit for rapid and reliable bisulfite treatment and conversion of DNA to achieve high-throughput methylation analysis. The key to the magnetic bead-based bisulfite conversion kits workflow is the convenient magnetic binding beads and the ready-to-use Lightning Conversion Reagent. Desulphonation and clean-up of the converted DNA is performed while bound to the MagBinding Beads. Whether using MagBead or spin-column cleanups, the high yield, converted DNA from these bisulfite conversion kits is ideal for PCR, array, and next-generation sequencing such as bisulfite sequencing, etc.
Comparison Table
| Catalog # | Product | Format | Elution Volume |
| D5049 | EZ DNA Methylation-Lightning Automation Kit | Magnetic Beads | ≥ 25 µl |
| D5030 | EZ DNA Methylation-Lightning Kit | Spin-Column | ≥ 10 ul |
| D5032 | EZ-96 DNA Methylation-Lightning Kit (Shallow-Well) | 96-Well (Shallow) | ≥ 30 µl |
| D5033 | EZ-96 DNA Methylation-Lightning Kit (Deep-Well) | 96-Well (Deep) | ≥ 15 µl |
| D5046 | EZ-96 DNA Methylation-Lightning MagPrep | Magnetic Beads | ≥ 25 µl |