Successfully Added to Cart
Customers also bought...
-
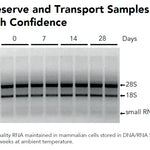 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
- No bisulfite conversion needed: from DNA sample to results in just one step.
- Accurate quantification of DNA methylation levels at the genomic regions of choice.
- Ideal for rapid screening of DNA methylation at single and multi-locus.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- No bisulfite conversion needed: from DNA sample to results in just one step.
- Accurate quantification of DNA methylation levels at the genomic regions of choice.
- Ideal for rapid screening of DNA methylation at single and multi-locus.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Description
Performance
Technical Specifications
| Compatible Thermal Cyclers | Bio-rad CFX-96™, CFX Opus 96™, QuantStudio qPCR Systems, Roche LightCycler 480®, or similar. |
|---|---|
| Detection Dye | Provided dye is compatible with SYBR Green Filter. Assay is flexible with other dyes and TaqMan probes. |
| Input | High quality DNA, suitable for use in restriction enzyme digestion. A260/280 > 1.8, A260/230 > 1.8. Optimal DNA input is 20 ng in 5 µl. Range validated 5 ng – 1 µg (in 5 µl). |
| Processing Time | Optimal 14 hours. |
| Storage | Store at -20°C for up to 12 months. Avoid repeated freeze-thaw cycles of reagents. Prolong storage at -80°C. 2X Test PreMix and 2X Reference PreMix are light sensitive. Minimize light exposure. |
Resources
Documents
FAQ
Yes, researchers may use alternative fluorescent dyes or molecular probes for Real-Time PCR. However, they must ensure that the settings on the Real-Time PCR instrument are adjusted to be compatible with the selected dye.
In principle, the OneStep Plus qMethyl PCR Kit can be applied to genomic DNA from any species, provided the genomic DNA is pure. Aim for 260/230 ratios of 2.0 to 2.2 and 260/280 ratios of approximately 1.8. Impure DNA can be cleaned up using Zymo’s DNA Clean & Concentrator-5 (D4013).
For this technology to perform optimally, amplicon must be bigger than 150bp. So, for example when using cfDNA, we would recommend using an input of 50ng or more for improved methylation assessment.
We recommend quantifying with Qubit.
Primers of the target of interest can be designed using conventional methods. We recommend carrying out a titration to determine optimal annealing temperature of the primers you have designed. We have validated the qmethyl I & II primers to work with annealing temperatures ranging from 56 - 64°C with 58°C being the optimal annealing temperature.
The MSRE digestion step - for 12 hours at 30°C – can be carried out in an incubator. We have validated this digestion step for the range of 2 – 24 hrs. The plate should then be transferred to a Real-Time PCR instrument for the remaining PCR steps. Optimal digestion time is 12 hours.
Yes, QuantStudio has been validated for the OneStep PLUS qMethyl PCR protocol with ROX turned off.
Yes, the protocol has been implemented in multiplexing with the use of dual-labeled TaqMan Probes. Taqman probed allow for testing of multiple targets in the same reaction. For further information, please contact Tech Support.
Need help? Contact Us