Microbiomics: Macro-Bias
Measurements from leaders in the field do not agree…
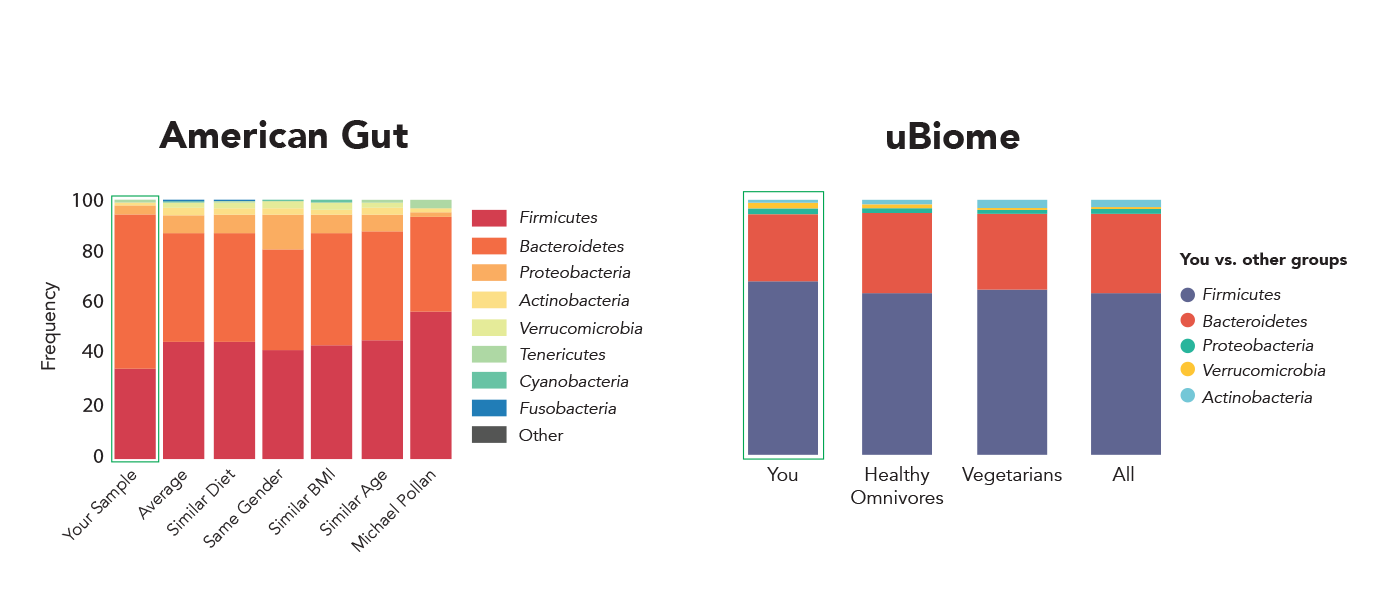
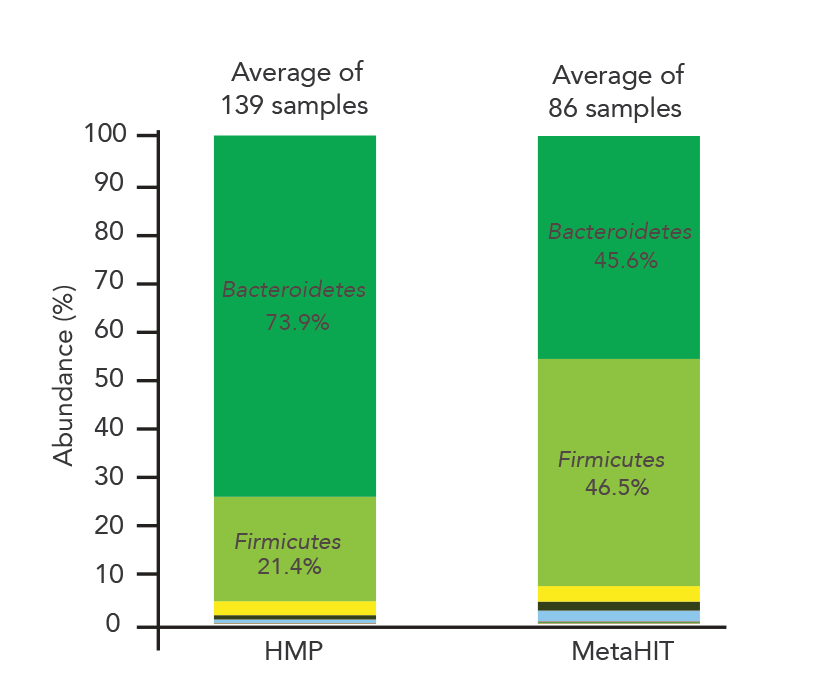
If you send the same sample to two different labs for analysis, you would expect the results to agree. Unfortunately, this hasn’t been the case for microbiome measurements even from well-known organizations. American Gut and uBiome are two of the best-known microbiome profiling organizations. But, given the same fecal sample, the two organizations have reported dramatically different microbial profiles (Figure 1) [1]. Even more strikingly, the two biggest microbiome profiling projects, Human Microbiome Project (HMP) and Metagenomics of Human Intestinal Tract (MetaHIT), analyzed hundreds of human gut microbiome samples and found the two dominant phyla (Bacteroidetes and Firmicutes) in dramatically different abundance (Figure 2). Unfortunately, this difference is likely due to technical variations in DNA extraction procedures used by the two projects as Wesolowska-Andersen et al. [2] pointed out.
What could go wrong?
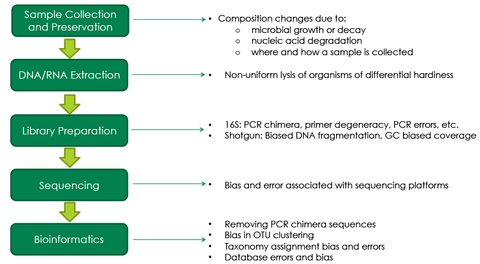
The contradictory results mentioned above are most likely due to bias generated due to the complex nature of microbiome measurements. An NGS microbiomics workflow has multiple steps and each step has various factors that can ause bias in the final result (Figure 3). Most protocols currently used in microbiomics experiments were designed before the development of the microbiomics field; therefore, they were not designed with the intricacies of bias in mind. For instance, although a conventional DNA extraction method have generated highly pure DNA, it may only extract DNA only from easy-to-lyse bacteria while leaving tough-to-lyse bacterial DNA behind leading to a substantially inaccurate profile.
What can be done?
Microbiome measurements demand more rigorous methods that are certified unbiased and low bioburden. To help researchers produce accurate microbiome measurements, Zymo Research is leading the development of new Microbiome-Grade tools for this field. These include the first commercially available microbiome reference materials, the first DNA extraction kit designed to ensure non-biased cell lysis, and the first cold-free and unbiased method to preserve microbial samples.
Learn More about the Microbiome-Grade Tools that Set the Standard for Accurate Microbiome Profiling:
References:
1. Saey TH: Here is the poop on getting your gut microbiome analyzed. In: Science News. vol. 2017; 2014.
2. Wesolowska-Andersen A, Bahl MI, Carvalho V, Kristiansen K, Sicheritz-Ponten T, Gupta R, Licht TR: Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014, 2:19.


