
Total RNA Sequencing
Compatible with low input, FFPE samples, and any organism (mammals, plants, bacteria, and more)
Validated solution for unbiased gene expression profiles
Comprehensive publication-ready bioinformatics gene expression analysis report
User Friendly & Comprehensive Report
Publication-Ready Gene Expression Analysis at the Touch of a Button

Full Analysis Includes:
- Sample Distance Matrix (PCA/heatmap)
- Differential gene expression
- Scatter plot
- MA plot
- Gene set enrichment
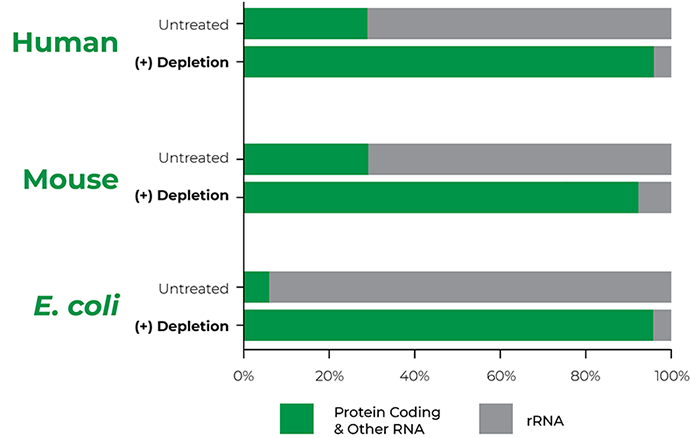
Unbiased Gene Expression Data with Probe-Free Technology
Workflow Certified Using RNA Standards of Known Composition
Probe-Free rRNA Depletion
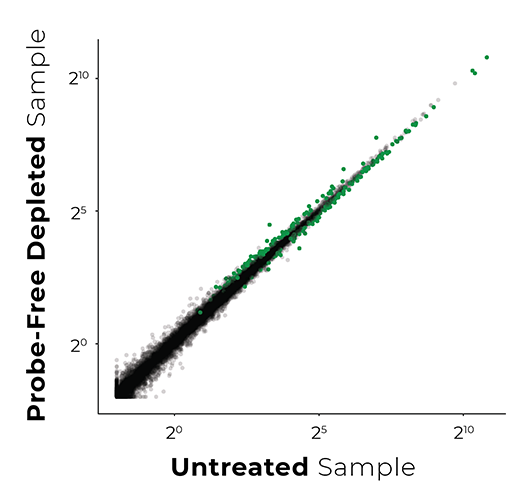
Unbiased Libraries
(264 genes affected)
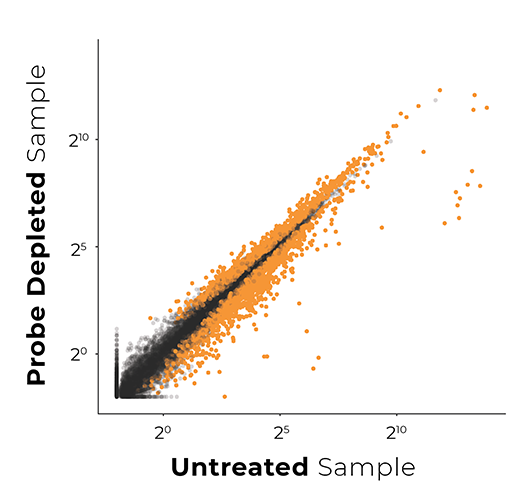
Probe-Based rRNA Removal
Biased Libraries
(3,603 genes affected)
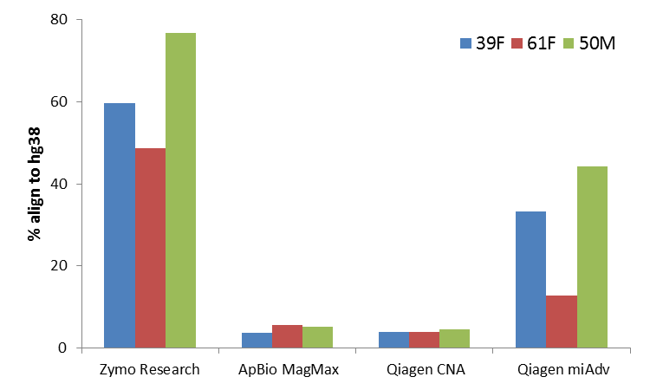
Robust Workflow Compatible with Any Sample Type
microRNA Sequencing
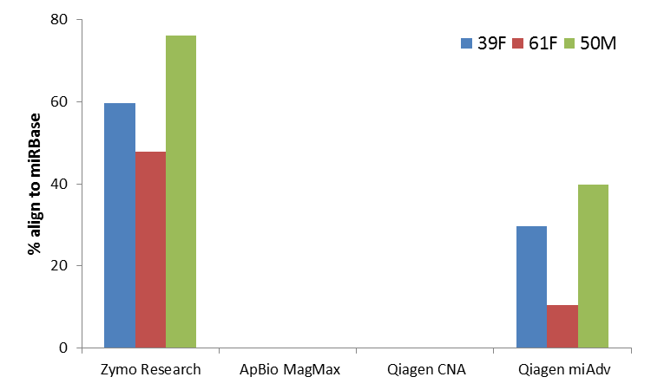
Efficient microRNA capture and isolation
Compatible with cfRNA from plasma and serum samples
Comprehensive publication-ready microRNA expression data analysis
Efficient microRNA Capture from Biological Samples
Total RNA-Seq Technical Specifications
| Sequencing Platform | NovaSeq 6000™ or NovaSeq X™ |
|---|---|
| Read Length | 2 x 150 bp, Unique Dual Indexing (UDI) |
| Sequencing Depth | ~30 mln read pairs (can be customized) |
| Accepted Sample Types | RNA, Cells, Tissue, FFPE Tissue Samples |
| Library Prep | Zymo-Seq RiboFree Total RNA Library Kit (cat. R3000) |
| rRNA Depletion | Enzymatic rRNA depletion compatible with any species |
| Data Visualization | Sample similarity matrix, MDS plot, heatmap with top gene expression patterns, scatter plot, MA plot |
| Statistical Analysis | Differential gene expression analysis with DESeq2 package, gene set entrichment analysis (if pathway annotation is available) |
Tell Us About Your RNA-Seq Service Project
Please describe your project and one of our scientists will reach out to assist you.

