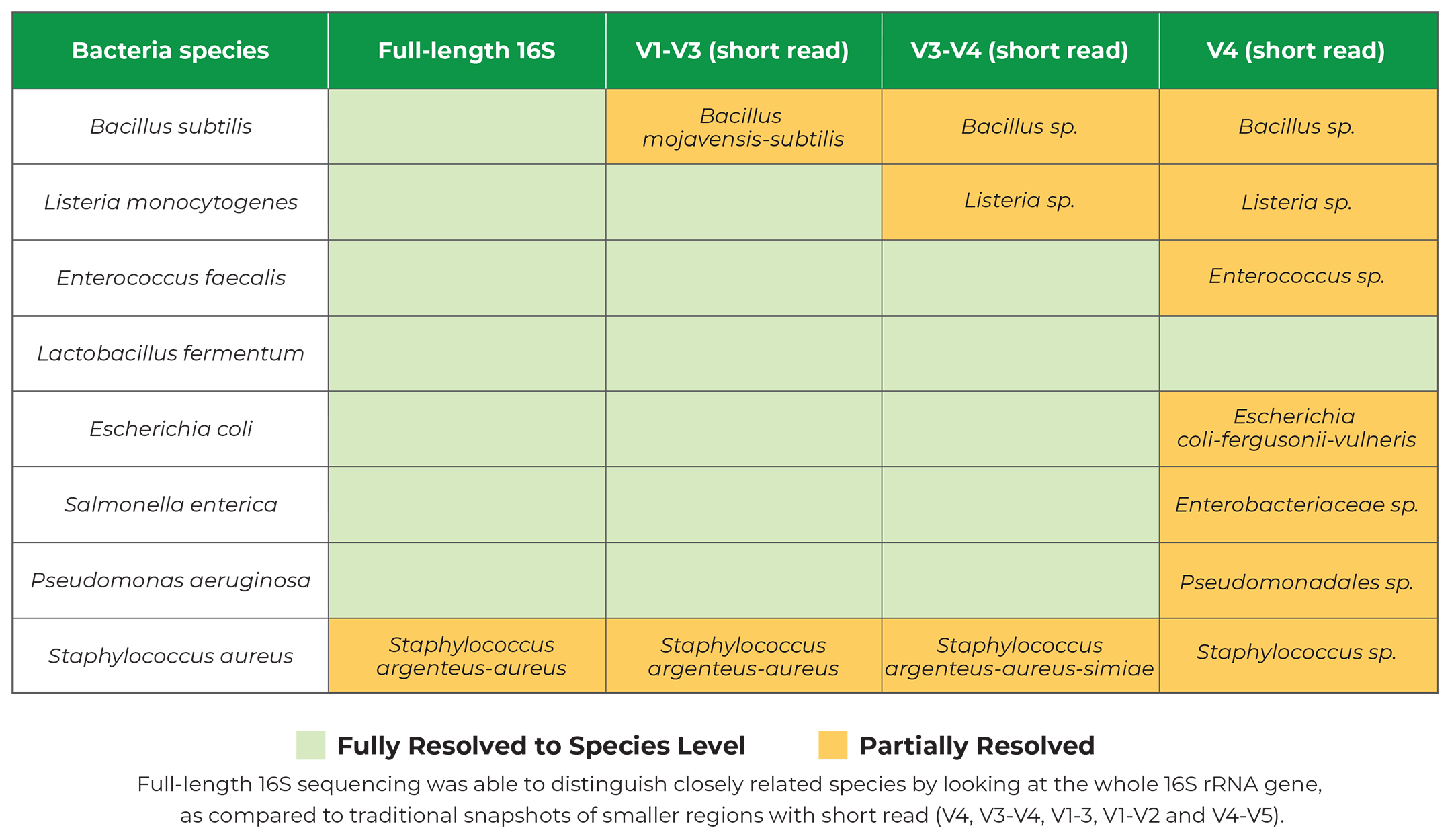
Targeted sequencing amplifying certain variable regions of 16S rRNA genes has become a popular and routine technique for
microbiome composition profiling. Traditionally, researchers relied on short-read sequencing platforms (e.g. Illumina
MiSeq) for this application. However, short-read sequencing read-length restricts this application to one or a few
variable regions of 16S rRNA genes, e.g. V4, V3-V4, V1-V3, V1-V2 and V4-V5. As a result, the taxonomy resolution is
limited.
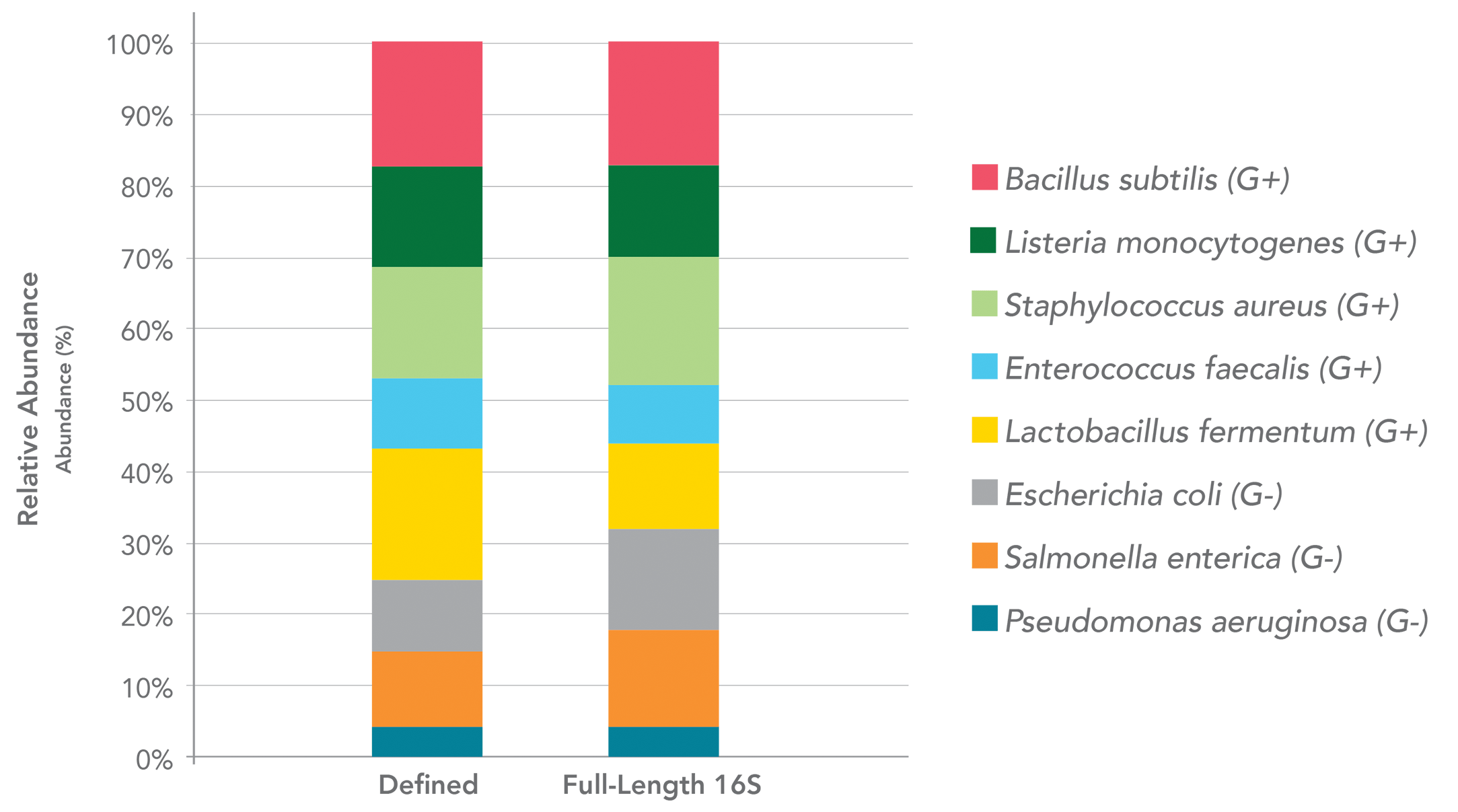
With the introduction of long-read sequencing platforms (e.g PacBio Sequel II and Oxford nanopore sequencing),
researchers can now leverage the complete taxonomy resolution power of the full 16S rRNA gene. Full-Length 16S
Sequencing is the latest in this long-read technology.
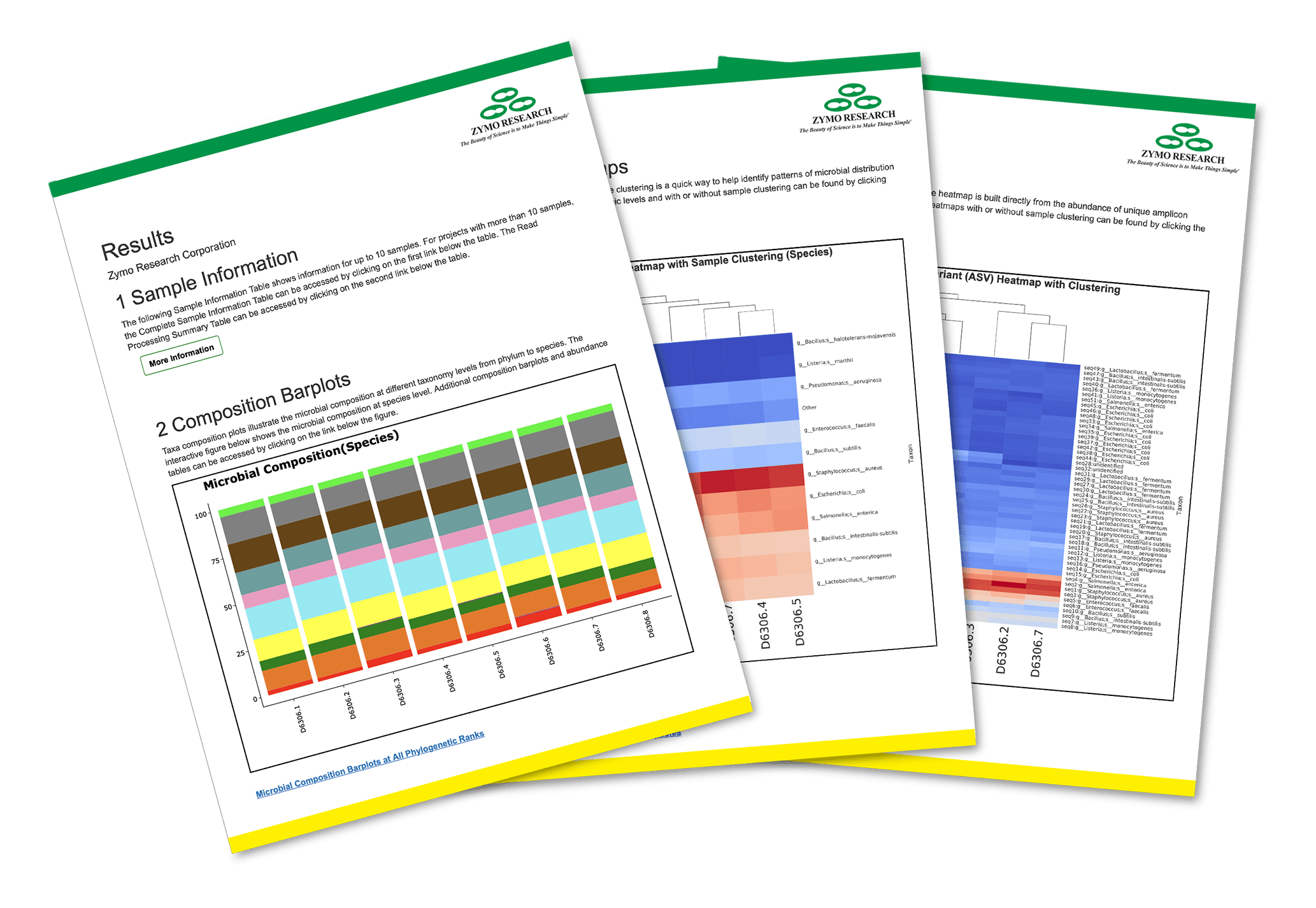
Zymo Research is a frontier in microbiome analysis and is one of the few Full-Length 16S Sequencing service providers.
In light of our motto to “Make Science Simple”, we are happy to be on the forefront of Full-Length 16S Sequencing
customer offerings. Zymo Research is happy to combine PacBio HiFi sequencing and error correction bioinformatics
analysis (e.g. Dada2), so that we can recover the error-free full-length 16S sequences from a mixed microbial community
(e.g soil or feces). We are offering this powerful Full-Length 16S Sequencing service for microbiome researchers and
industry partners alike. As a Certified PacBio Provider you can rest assured that your samples will be well taken care
of as they undergo Full-Length 16S Sequencing.