The Congress shall have Power… To coin Money, regulate the Value thereof, and of foreign Coin, and fix the Standard
of Weights and Measures
–U.S. Constitution, Article I, Section 8, Clause 5
While units such as the mile, the kilogram, the watt, and the calorie are thrown about in casual conversation without much consideration to their importance in our modern world, the Framers of the U.S. Constitution thought these units of weight and measure so critical that authority over them was granted in the same breath as that over money. One of the earliest measurements that needed to be taken with some precision was length, as humans began to discover that the tools we discovered could be used to produce buildings, and those buildings were much more robust if all of the support structures were the same height.
Early measurements of length might have been based upon the length of someone’s forearm (called a cubit), their foot, or the width of their thumb. While this was a unit of measurement, variability between individuals made standardization difficult and led to issues where precision was necessary. The solution to this came when some individual took a nearby wooden rod and cut it to some specific, convenient length and added notches to the rod with their knife to indicate smaller measures This first rod would have been one of the earliest standards and anything measured against that standard rod or rods made from it would be that much more precise.
When diving into the world of microbiome research, precision and reliability are everything. In our parent blog, How to Master the Microbiome, we explored the key steps to unlocking the secrets of microbial communities—from sample collection and preservation to sequencing and bioinformatic analysis. A critical component that underpins the success of each of these steps is the use of microbiome standards and controls. These tools ensure data accuracy, reproducibility, and confidence in your findings, helping you navigate the challenges of microbiome studies with ease. In this post, we’ll dive deeper into why microbiome standards and controls matter, how to use them effectively, and what innovations are shaping the field.
The Need for Microbiome Standards: Ensuring Accuracy and Reproducibility
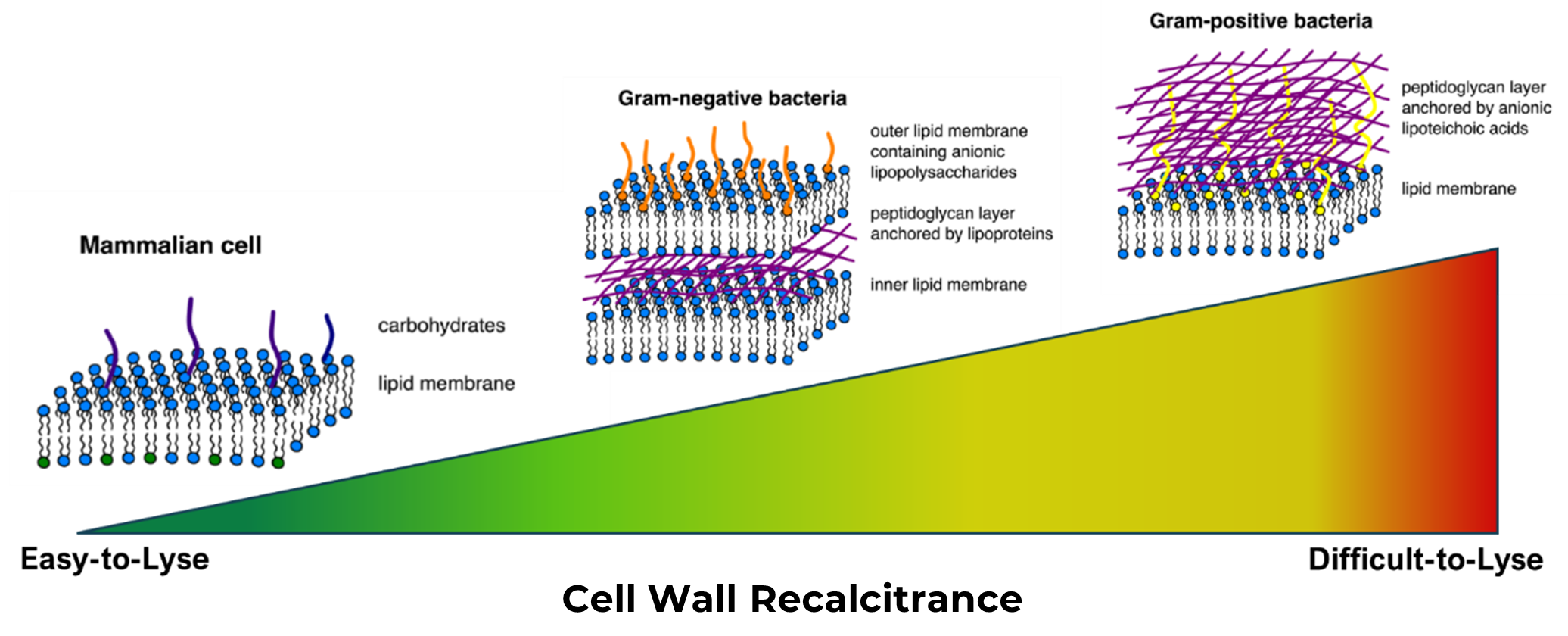
Microbiome research relies heavily on high-throughput sequencing, also known as Next-Generation Sequencing (NGS), a powerful technique that enables in-depth community profiling of microbial populations. Despite its potential, microbiome workflows are fraught with biases and inconsistencies that can skew results. One significant source of bias in microbiome studies is the DNA extraction process. This so-called "lysis bias" refers to the differential efficiency with which microbial cells are broken open to release their DNA. Some microbes may be lysed more readily than others, leading to an inaccurate representation of the microbial community. How easy or difficult a cell is to lyse is a direct function of how extensive its cell wall structure is built. For example, a mammalian cell with only a lipid membrane to maintain the integrity of the cell will lyse quite readily. Conversely, Gram-positive bacteria, with their elaborate peptidoglycan cell walls, will require significantly more effort in order to disrupt. At Zymo Research, we like to refer to this particular characteristic of cells as “cell wall recalcitrance,” with those having a higher cell wall recalcitrance being increasingly more difficult to lyse. Below is a visual representation of the spectrum of cell wall recalcitrance, with examples of where various cell types can be expected to fall:
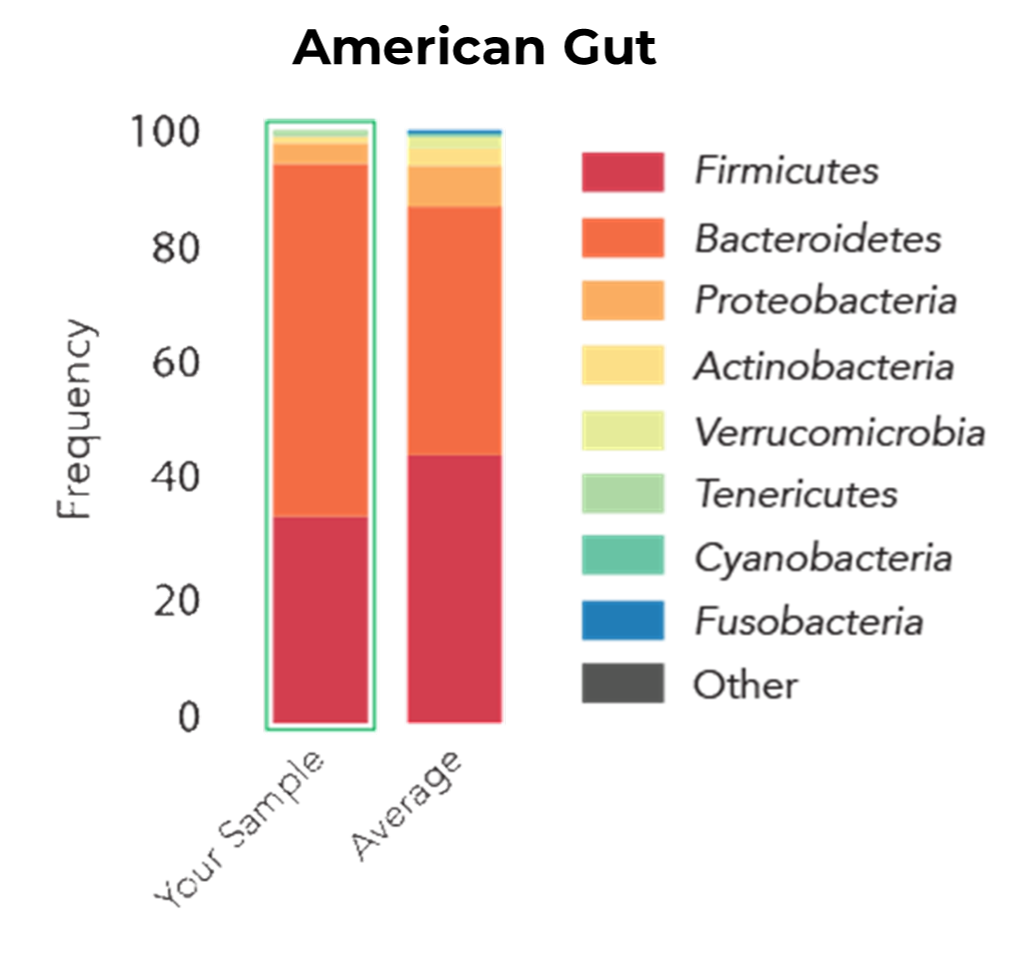
A high-profile example of bias in action can be found in the 2014 Science News article “Here's the Poop on Getting Your Gut Microbiome Analyzed,” wherein the same fecal sample was submitted to two different gut microbiome analysis services. The author found that the microbial profile of the sample differed greatly, depending on which service performed the analysis. In the results provided by American Gut, Bacteroidetes – a phylum of Gram-negative bacteria – were the dominant taxonomic group for both the author’s sample and the aggregate. In contrast, µBiome’s analysis instead suggests that Firmicutes – a phylum of Gram-positive bacteria – were the dominant taxonomic group in both cases. Given the nature of the discrepancy between these two results, one possible explanation for the observed difference is suboptimal lysis methods used in the American Gut analysis, leading to an underrepresentation of Gram-positive bacteria (i.e. Firmicutes) present in the sample.
Even before sequencing, evidence of potential lysis issues may be detected. The expected yield of DNA from our mock community standard is 2 micrograms for 75 microliters of standard. If significantly less DNA is obtained, it is worth determining the reason for this discrepancy. If later sequencing reveals that along with low yield, there is significant bias showing an overabundance of gram-negative microbes, this is a strong hint that there is a lysis failure occurring that needs to be addressed. If the low yield is happening in the absence of any evidence of lysis bias, the DNA purification and recovery should be examined along with determining if the method of measurement is potentially able to explain the different observation. Additionally, a standard sample should be run on a regular basis—ideally with each sample prep run—and the yield measured to ensure consistency; if abnormally low yields are observed using a consistent measurement system, that is strong evidence that something has changed.
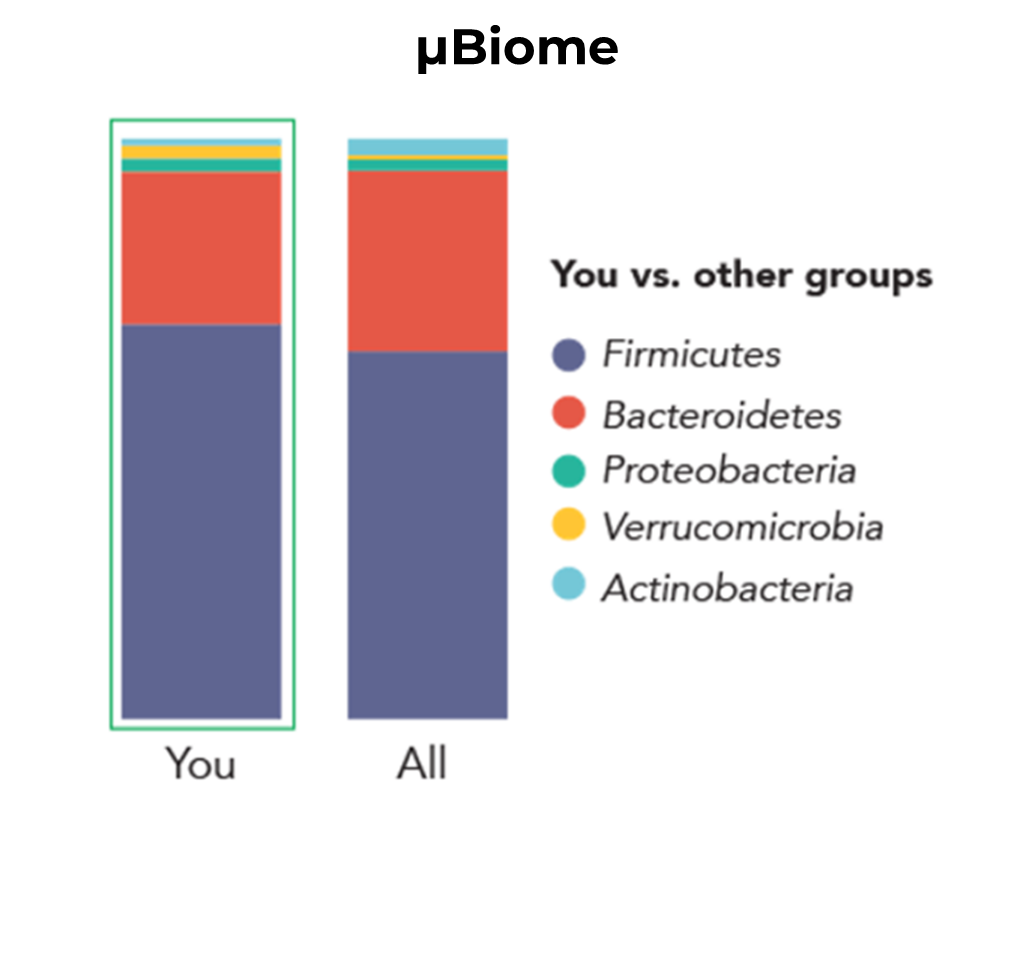
Another common source of bias, that is also capable of contributing to the discrepancy observed above, is amplification bias. Sometimes also referred to as G/C bias, amplification bias is the result of varying amounts of guanine and cytosine pairs in genomic DNA content. Because guanine and cytosine bond more strongly than adenine and thymine, DNA fragments containing more G/C pairs are more difficult to denature during PCR amplification. Therefore, NGS library prep protocols with suboptimal PCR conditions, or which require more extensive PCR amplification, can impart significant bias into the analysis. Firmicutes are typically classified as having relatively low percentages of genomic G/C content, so the proportionally higher representation of Firmicutes in the µBiome analysis could also be described as overrepresentation due to amplification bias. In truth, while we can speculate, it is impossible to say for certain what biases have been imparted on a sample such as this, and to what extent they have influenced the observed profile. However, by submitting the same fecal sample to both services, the author highlighted the need for standards and controls in microbiome research. By using a well-defined standard reference material for microbiome quality control, rather than a fecal sample of unknown composition, the amount and nature of bias can be determined, and addressed before any real samples are processed. The remainder of this blog will discuss types of standards that can be used as well as how to use them. Each of the upcoming sections will introduce a key concept related to the use of standards that will be expanded upon in its embedded video.
Key Terms and Concepts for Microbial Standards
The most basic term and concept to understand with respect to microbial standards is what defines a standard reference material. The National Institute for Standards and Technology (NIST), the US Federal Institute that oversees weights and measures and whose authority originates with the part of the Constitution quoted at the start of this writing, defines a reference material as, “Material, sufficiently homogenous and stable with respect to one or more specified properties, which has been established to be fit for its intended use in a measurement process.” While this definition is extremely dense in meaning, a few critical pieces stick out:
-
Material: It must be some physical item. A digital standard can be standardized data, but not a reference material
-
Homogenous and stable: The material properties should be consistent throughout and should not change significantly when stored for a specified amount of time
-
One or more specified properties, fit for its intended use: There are one or more properties for which the material is intended as a reference measurement. There will likely be other properties that can be measured, but the material may not be a reliable reference for those.
Microbiome reference materials may come in different forms with different properties with respect to measurement and, while they all will contain some amount of microbial material, they will also likely contain other material as well. This other material is known as the matrix and it can be as simple as a microbial preservative to ensure shelf stability or as complex as fecal matter or tissue. Additionally, microbiome references can be known input or complex/natural with the former generally being pure, cultured microbial strains combined in a laboratory setting where the composition can be truly known and the latter being material derived from nature where an exact composition may not be knowable.
Some reference materials may be able to be added to real samples as a spike-in whose reads or other signal can be separated from the original sample and can then be used for comparison purposes on the individual sample. Any sample should have some known degree of manufacturing tolerance that can describe properties such as how far the actual material in the tube may deviate from the expected or how much variability there may be aliquot-to-aliquot.
With respect to measurements, quantification may be relative, indicating that results will be comparable to the reference material or absolute indicating that some exact concentration or count may be derived from the measurement. These measurements are often bioinformatic in nature and the bioinformatics involved can be generally divided into taxonomic assignment or attempting to place the read somewhere within the entire tree of life (or some portion of it) and reference alignment or trying to place the read somewhere within an expected genome or set of genomes provided to the aligner.
Microbial Tools of the Trade: Types of Microbial Standards
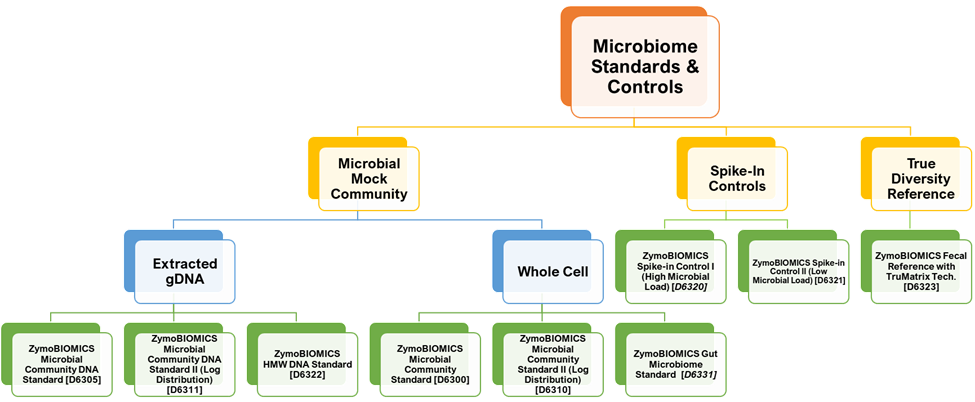
There are several types of microbial standards offered by Zymo Research and other entities, and selecting the correct standard can be daunting even for experts, let alone newcomers to the field. Often, a few questions can be used to suggest the appropriate reference material or understand how a given standard reference material may be used:
-
Known-input or complex material?
-
A known input material will often be simpler to analyze and because it was generated in a laboratory, it is very likely that there will be more precise expected values to allow for better gauging of result accuracy.
-
A complex material is likely to require a more complex analysis and will usually have fewer (if any) expected values reported, with those reported often being less precise. A complex material does have the potential to mimic a real sample matrix from nature, which is difficult to achieve in known-input. Additionally, a complex material can meet or exceed the microbial complexity of a real sample, providing a more realistic benchmark and possible stress-test for bioinformatic or other analytic pipelines.
-
-
Spike-in or stand-alone sample?
-
A spike in can go into the sample itself and serve as a marker for different aspects of the process (such as verifying unbiased lysis of the individual sample). Additionally, if an absolutely known quantity is spiked in, the reference material can be used as a “genomic yardstick” to compare and quantify what is detected. A spike-in standard can potentially have its signal or reads confused with those from the sample itself, potentially causing confusion and making the sample unusable. This risk is greater if the spike-in is selected without consideration for what may be present in the sample.
-
Stand-alone reference materials are run as their own sample and are not added to real samples for analysis. Stand-alone references can be run through the same process to assess quality (such as lysis efficiency) and can have more microbes added with those microbes potentially being expected in the real samples as well.
-
-
Whole-cell or DNA
-
A whole-cell standard will require lysis in order to extract DNA. This provides an ability to evaluate lysis and extraction processes and determine if significant bias was introduced. If evaluation of upstream processes like lysis and extraction are not desired, this introduces a potential new variable into the mix.
-
Genomic DNA standards have purified DNA from the microbes used in the standard and require no lysis. Their use can identify biases that happen downstream of lysis and extraction, such as G/C biases in library prep or abnormal behaviors in the bioinformatic pipelines. This kind of standard removes any confounding factors that can arise during lysis and extraction.
-
Questions to Ask When Setting up a Pipeline
When initially establishing a microbiome workflow, standards can be used to establish the precision and accuracy of the outputs and optimize different components to maximize these attributes. Because a known-input standard can be run through as a sample and the results can be compared to determine how well the expected results were captured, this kind of standard provides an excellent starting point for workflow evaluation. Paired with a bioinformatics package such as the MIQ Score to provide a metric of deviation from expected results, accuracy of a pipeline can be easily determined even without significant computational biology experience. Taking this a step further, one can ask how consistent their procedures are by repeatedly running standards on different runs and days, with different team members performing the work, and using different pieces of equipment that will be available to the workflow. Ideally consistently high accuracy should be observed with as little variance as possible; high variance in accuracy scores may suggest some inconsistent practice in the workflow. Finally, different pieces of equipment can be evaluated to determine their effects on the accuracy of the workflow. An example of this would be testing out different bead lysis methods with multiple replicates and determining which provides the most consistent and best accuracy of those tested.
Questions to Ask When Reviewing an Established Workflow
We’ve all been there before, the results are back and they just don’t seem to match with what we were expecting or sometimes don’t even make sense. This is when an established workflow needs to be evaluated and reasons that might explain why such strange results were observed need to be isolated and uncovered. If robust microbiome quality control practices were in place and testing against a reference standard was being carried out regularly, this investigative process can be initiated quickly if deviations from expected performance is detected.
Being able to isolate different components of the workflow can make uncovering deficiencies much easier and a more methodical process. A great place to start this process is with a known-input standard using both a whole-cell and genomic DNA material. The whole-cell standard can be introduced as a sample at the start of the processing while the genomic DNA standard can be introduced after the lysis and extraction process. Any deficiencies arising lysis and extraction will not be present in the genomic DNA, and by comparing the performance on genomic DNA and whole-cell standards, it is possible to determine how much inaccuracy is being contributed by the lysis and extraction process vs. any downstream processing. In the next section, we will talk about how to isolate deficiencies arising from bioinformatics from those arising from upstream processes.
In any microbial community of interest, there are often microbes that are more desirable and those that are less desirable or sometimes even harmful. Often, development of microbiome-based applications centers on increasing the presence of specific microbes or decreasing the presence of others. If the only metric being taken is relative abundance, it can be easy to think that the presence of the desired microbe has been significantly increased when in reality a large portion of the community has been removed and the desired microbe is present in no greater absolute amount than before with everything else just diminished. This is where a spike-in reference material can be added to the sample before processing. The reads from the spike-in can be bioinformatically separated (assuming an appropriate spike-in was used) and then their quantity used as a “molecular yardstick” to compare absolute abundances between samples.
Finally, many modern studies are run out of multiple centers and have several individuals processing samples, often on different pieces of equipment. It is critical that reproducibility between sites and individual sample handlers be tested and maintained. While known-input materials can be useful for initial evaluations, a complex reference material, such as the Fecal Reference with TruMatrix Technology can provide a more “real” sample and matrix to test against. The results between different preparations can be compared, and while there is no “ground truth” expectation for this material, the degree of similarity between different runs can be used to quantify how repeatable results are between sites, teams, or machines.
How to Think About Bioinformatic Analysis
While bioinformatic analysis is a complex topic that could easily be its own blog, one of the most important concepts to understand when using reference materials is how to look at the results of an alignment-based analysis vs. a taxonomic assignment. As mentioned above, alignment starts with a limited set of known, expected genomes for the sample. This is not something that can be done for most microbiome samples, as it would create a massive reference bias and it is rare to have known, expected microbes with high-quality genomes prior to analyzing the sample. Alignment of reads is computationally a much simpler process than taxonomic assignment of reads. While counterintuitive, this is often the better choice for beginning to analyze data from a known-input standard. Using a known-input standard, we have much more prior knowledge of the sample than we would for nearly any other sample and can leverage that information, essentially saying that any read that can be reasonably explained by an expected microbe from the sample should be explained by that microbe, regardless of other microbes not in the standard that could also explain it. This removes a significant amount of uncertainty when quantifying microbes to determine accuracy relative to our expectations for the known-input standard.
Reanalyzing the known-input standard reads using a taxonomic assignment pipeline after the initial analysis using alignment can be highly informative. Taxonomic assignment pipelines vary in their exact algorithm, but all of them require a reference database containing the genomes of many organisms in a defined tree-of-life structure. By tracking where results for individual reads are either concordant (reads are attributed identically or very similarly), mostly concordant (reads are not attributed very closely, but not in a manner that disagrees), and discordant (read attributions disagree between methods). Identifying scenarios where there is a high count of discordant reads between methods and tracing the source can help identify potential weaknesses in the taxonomic assignment method or the reference database being used. An example of this would be reads aligning to a specific firmicute being taxonomically assigned to a fungus; these would clearly be discordant reads and should be investigated. Depending upon the results of the investigation, it could be discovered that a firmicute is inappropriately entered in the reference database in this scenario or that the taxonomic assignment algorithm is prone to errors in this scenario.
Starting with Standards in Your Workflow
While there are many standards available, just beginning with the ZymoBIOMICS® mock microbial community as either a whole-cell standard, genomic DNA standard, or both is a great place to start. In addition to the material, Zymo Research also provides an automated bioinformatic analysis tool in the form of the MIQ Score, which only needs reads and some information on your library prep to determine accuracy and generate a report that both quantifies accuracy and creates figures that may help explain any loss of accuracy based upon known bias patterns (such as lysis or GC-content).
If you would like to get started, please reach out to our microbiomics team for any advice you may need as well as to place your first order or request a sample of our mock microbial community standard to get started. While you’re here, please check out some of our other blog posts that may help your in your journey to developing a better microbiome pipeline.
