The Microbiomics Enlightenment
“Isn’t it possible that your results are due to an artifact?”
It’s a simple question that makes some microbiomics researchers break into a cold sweat. The excitement of exploring the world’s microbiomes and the ever-increasing availability of NGS, created a rush in microbial profiling studies that boomed at breakneck speed. But recently, many scientists have stopped to question the accuracy of such studies since many did not incorporate well-defined and validated controls. Without a positive control, such as a mock microbial community to emulate a diverse collection of microbes, there is no way of eliminating artifacts as possible factors in the results.
It is understandable why these controls were not used in the early days of microbiomics because well-characterized mock microbial communities are difficult to produce and thus, biases inherent to older nucleic acid extraction technologies were not yet realized until challenged with such controls.
It’s pronounced “Jim Bee”
Today, many researchers and some newly formed initiatives are pushing for the standardized use of such controls in all metagenomics studies, as spotlighted in this recent Technology Feature from Nature Methods1. One group tackling these problems is Stanford University’s Joint Initiative for Metrology in Biology (JIMB, pronounced “Jim Bee”), which was born out of the National Institute of Standards and Technology (NIST). One of the goals of JIMB is to promote and facilitate the adoption of standards and controls in metagenomics.
Making Standards a Standard
One commercially available source of microbiome standards that address these needs is Zymo Research’s ZymoBIOMICS™ line of standards and controls. These include mock communities in even-distribution and log-distribution formats to test extraction and library preparation methodology, as well as a spike-in control for absolute quantification of microbes by NGS measurements.
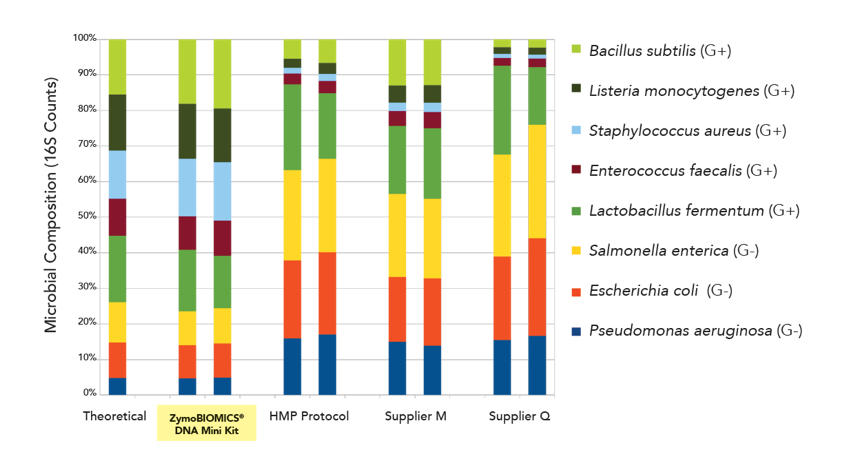
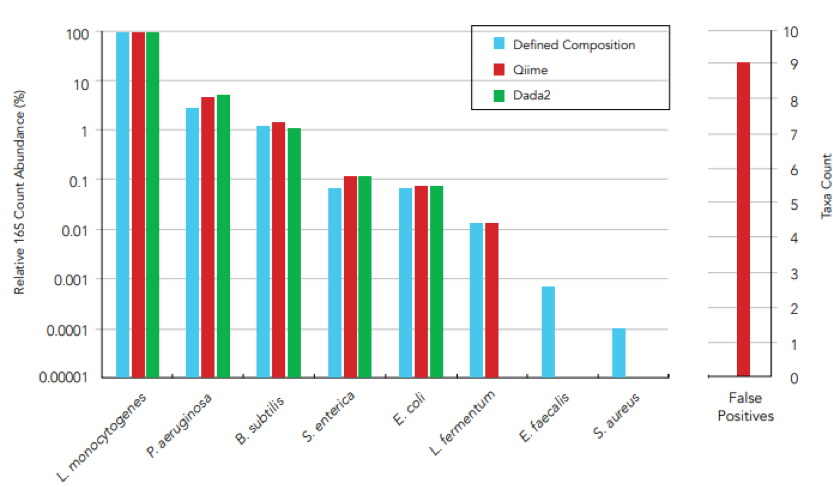
They consist of a combination of easy-to-lyse and tough-to-lyse organisms that allow for the validation of DNA extraction methods (figure 1) and with a varying level of genomic GC content for challenging library preparation and sequencing workflows (figure 2). The precision with which these standards are produced and validated is critical to their use as accurate metagenomics controls.
This well-defined composition is the reason these standards have been used in many high-impact studies including: Chris Mason’s comprehensive benchmarking study2, Rob Knight’s high-throughput 16S library prep workflow3, the Loman Lab’s long-read sequencing project4, and many more.
A Bright Future for Experimental Controls
Due to the lack of validated, high quality mock microbial communities in the early days of microbiomics, it is understandable why many studies did not incorporate controls in their experiments. However, recent years have seen a sort of enlightenment in microbiomics, in which researchers are seeking microbiomics-grade, non-biased extraction methods as well as accurately defined mock microbial communities. With the push for updated methods and controls being spearheaded by field leaders as well as distinguished consortia such as NIST and JIMB, we are certainly on the precipice of a golden age for accurate microbiomics breakthroughs.
References:
1. Marx V. Controls let genomics experimenters drive with a dashboard. Nature Methods. 2019 16(1): 29-32.
2. McIntyre ABR, Qunit R, Afshinnekoo E, Prill RJ, Alexander N, Minto SS, Danko D, Foox J, Ahsanuddin S, Tighe S, Hasan NA, Subramanian P, Moffat K, Levy S, Lonardi S, Greenfield N, Colwell RR, Rosen GL, Mason CE. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biology. 2017 18(1): 182.
3. Minich JJ, Humphrey G, Benitez RAS, Sanders J, Swafford A, Allen EE, Knight R. High-throughput Miniaturized 16S rRNA Amplicon Library Preparation Reduces Costs while Preserving Microbiome Integrity. mSystems. 2018 3(6): e00166-18.
4. Nicholls SM, Quick JC, Tang S, Loman NJ. Ultra-deep, long-read nanopore sequencing of mock microbial community standards


