Successfully Added to Cart
Customers also bought...
-
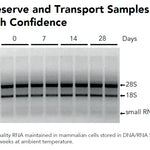 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
- Pellet-Free: Direct lysis procedure omits cell-pelleting and resuspension steps.
- High Quality: Ready for PCR, sequencing, cloning, and transfection.
- Streamlined Protocol: Culture, lyse, and neutralize in the same plate, saving time and reducing plastic waste.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Pellet-Free: Direct lysis procedure omits cell-pelleting and resuspension steps.
- High Quality: Ready for PCR, sequencing, cloning, and transfection.
- Streamlined Protocol: Culture, lyse, and neutralize in the same plate, saving time and reducing plastic waste.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | Price | |
|---|---|---|---|---|
| D4036-5-60 | Zyppy Elution Buffer | 60 ml | $87.00 | |
| D4041-1-30 | Deep Blue Lysis Buffer | 30 ml | $55.00 | |
| D4041-1-48 | Deep Blue Lysis Buffer | 48 ml | $83.00 | |
| D4041-4-100 | Neutralization/Clearing Buffer | 100 ml | $96.00 | |
| D4041-4-200 | Neutralization/Clearing Buffer | 200 ml | $165.00 | |
| D4036-5-30 | Zyppy Elution Buffer | 30 ml | $46.00 | |
| D4036-5-100 | Zyppy Elution Buffer | 100 ml | $108.00 | |
| D4036-4-48 | Zyppy Wash Buffer (Concentrate) | 48 ml | $59.00 | |
| D4036-4-24 | Zyppy Wash Buffer (Concentrate) | 24 ml | $41.00 | |
| D4036-3-160 | Endo-Wash Buffer | 160 ml | $67.00 | |
| D4036-3-120 | Endo-Wash Buffer | 120 ml | $59.00 | |
| C2007-24 | 96-Well Plate Cover Foil | 24 Foils | $46.00 | |
| C2007-6 | 96-Well Plate Cover Foil | 6 Foils | $13.00 | |
| C2011-8 | Air Permeable Sealing Cover | 8 Pack | $45.00 | |
| C2011-4 | Air Permeable Sealing Cover | 4 Pack | $25.00 | |
| C2011-2 | Air Permeable Sealing Cover | 2 Pack | $13.00 | |
| C2007-12 | 96-Well Plate Cover Foil | 12 Foils | $23.00 | |
| C2004 | Zymo-Spin I-96 Plate | 2 Plates | $171.00 | |
| P1001-2 | 96-Well Block | 2 Blocks | $22.00 | |
| C2002 | Collection Plate | 2 Plates | $26.00 | |
| C2003 | Elution Plate | 2 Plates | $23.00 | |
Description
Performance
Technical Specifications
| Applicable For | Ligation, sequencing, restriction endonuclease digestion, in vitro transcription, and other sensitive applications requiring pure DNA. |
|---|---|
| Elution Volume | ≥ 30 µl |
| Equipment | Centrifuge with microplate carriers with height tolerance of 60 mm (2.36 inches) |
| Processing Time | 45 min |
| Purity | Typical Abs 260/280 ≥ 1.8. |
| Size Range | Up to 25 kb |
| Yield | Up to 5 µg per preparation depending on the plasmid copy number, culture growth conditions, and strain of E. coli processed. |
Resources
Documents
FAQ
No
10 mM Tris-HCl, 0.1 mM EDTA, pH 8.5.
Yes, all our plasmid preps have an endo-wash step to remove endotoxins and produce plasmid DNA that is ready for transfection with most cell lines.
No, we recommend using our ZR BAC DNA Miniprep Kit or ZymoPURE II Kits for isolating large constructs.
≤ 50 Endotoxin Units/µg of plasmid DNA.
Yes, the RNase A is fairly stable at room temperature, but we recommend placing it in 4°C as soon as possible.
Zyppy Wash Buffer is available for purchase, separately. However, you can substitute with DNA Wash Buffer, Plasmid Wash Buffer, or 80% ethanol.
Yes, the RNase A is fairly stable at room temperature, but we recommend placing it in 4 °C as soon as possible to ensure optimal performance throughout the life span of the product.
No, the sediment inside the buffer is insoluble, but it needs to be completely resuspended in the buffer prior to use for optimal performance.
Citations
Need help? Contact Us




