How to Discover Biases in Metagenomic Studies
The growth of metagenomic studies has revolutionized our understanding of the relationships between microbiota and the environment or health.
While this realization has resulted in many new discoveries, data reproducibility has remained a challenge. This issue spans metagenomic research across labs and stems from the fact that bias can be introduced at various steps across the metagenomics workflow, as observed by many in the field 1-6.
The problem of bias is so widespread that even submitting the same sample to two different microbiome profiling organizations can yield results that are dramatically different from one another (Figure 1).
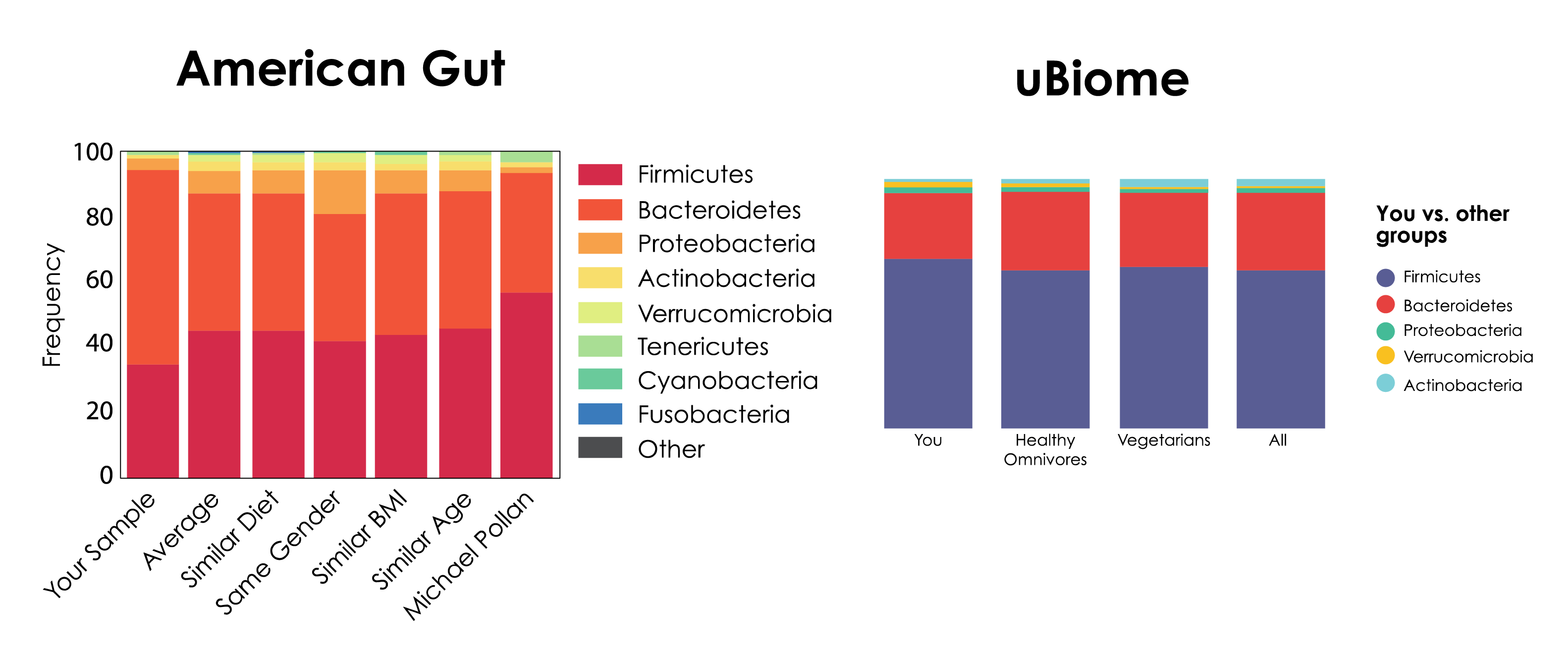
These biases can arise at every step throughout the entire metagenomics workflow. However, one of the most problematic steps that contributes to bias lies in nucleic acid extraction. With growing evidence of systemic biases, the need for more accurate metagenomic nucleic acid extraction workflows is now larger than ever.
How do Biases Occur Within Extraction?
Microbial communities are complex and diverse, consisting of Gram-positive bacteria, Gram-negative bacteria, and fungi. Accurate metagenome profiling requires the liberation of DNA from all the diverse species within a microbial community. However, it is common to observe ineffective lysis during the nucleic acid extraction which then leads to microbial profile bias. This is due to some microbes being very difficult to lyse 6, 8. If the cells are not lysed, the DNA will remain locked away within the cell and will not be purified or detected.
It has been shown that processes utilizing chemical or thermal lysis overrepresent the easy-to-lyse organisms (Gram-negative bacteria) due to this very reason. Since the tough-to-lyse organisms (e.g. Gram-positive bacteria and yeast) are more resistant to DNA liberation, it causes a bias towards the easy-to-lyse species. Many extraction protocols do not account for these vast differences in sample composition meaning it is common to observe non-uniform lysis and microbial profile bias 9.
Extraction protocols that utilize mechanical lysis (e.g. sonication, blending, liquid nitrogen/mortar and pestle, French pressing, and bead-beating) are considered the best approach to microbial lysis due to their stochastic nature with bead beating referred to as the gold standard. However, these mechanical lysis methods still need to be optimized or they will suffer from issues such as low yield, excessive nucleic acid shearing, non-uniform lysis, excessive heat, and shear forces.
How Can Bias Be Discovered?
The only true way to know if an extraction system is introducing bias into a metagenomic study is to evaluate the system with a microbial standard. A microbial standard refers to a pool of various microorganisms (including both Gram-positive and Gram-negative species) that act as a mock microbial community and mimics the metagenomic populations present within samples. This standard is processed normally through the extraction workflow.
Since the abundance of each microorganism in the microbial standard is known, the results obtained from the 16s sequencing data should match closely to the standard. Large deviations from this indicate that the extraction system introduced bias into the results. Most commonly, these deviations reveal themselves as an overrepresentation of Gram-negative species in the population. This can be seen clearly in a comparison of various extraction systems (Figure 2).
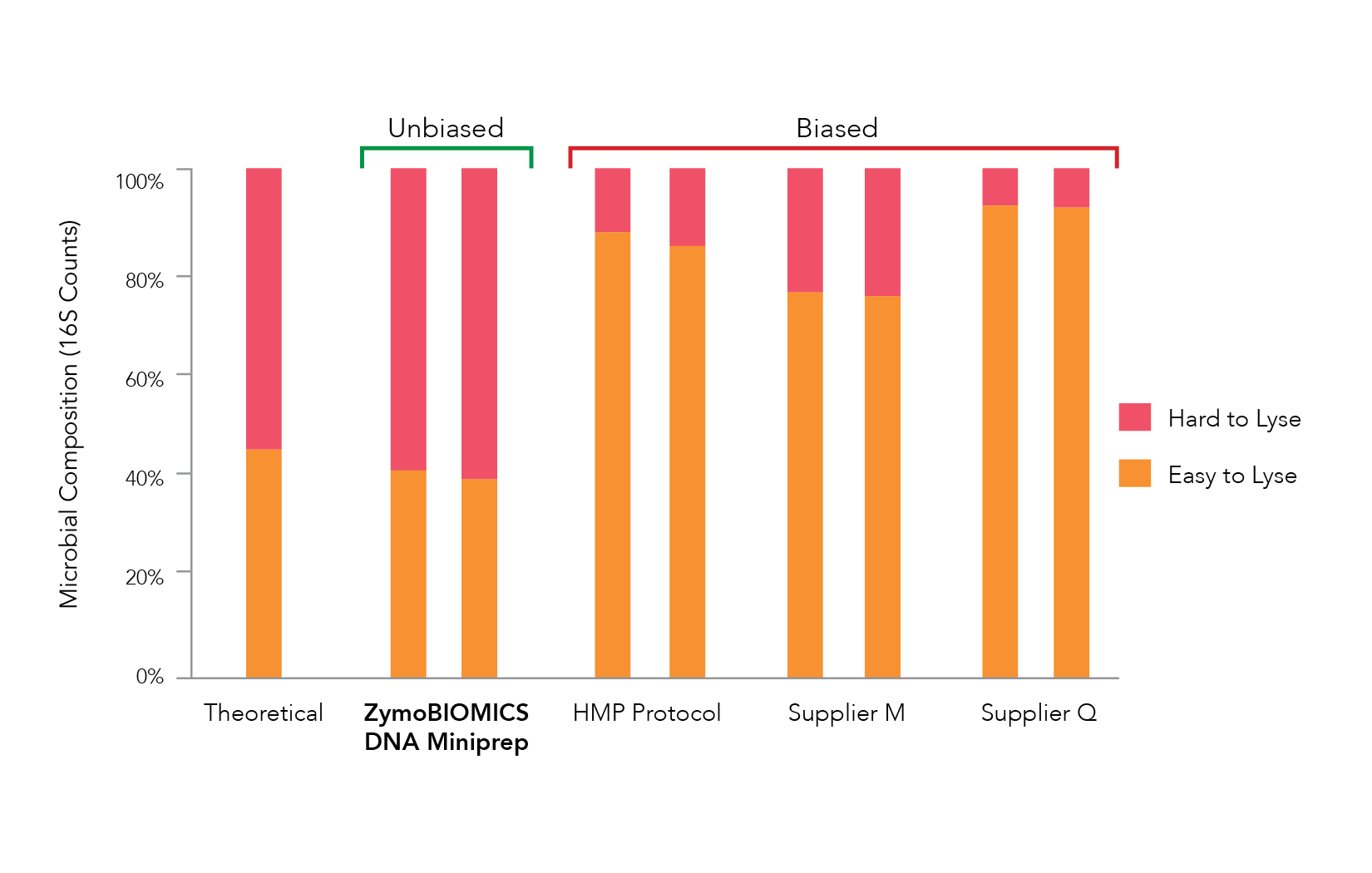
Bias-free Methods
The ZymoBIOMICS line addresses this key challenge of bias within a metagenomics workflow. The ZymoBIOMICS 96 Magbead DNA Kit utilizes mechanical lysis that has been developed and optimized with microbial community standards to ensure complete lysis of all the tough-to-lyse organisms (Figure 3).
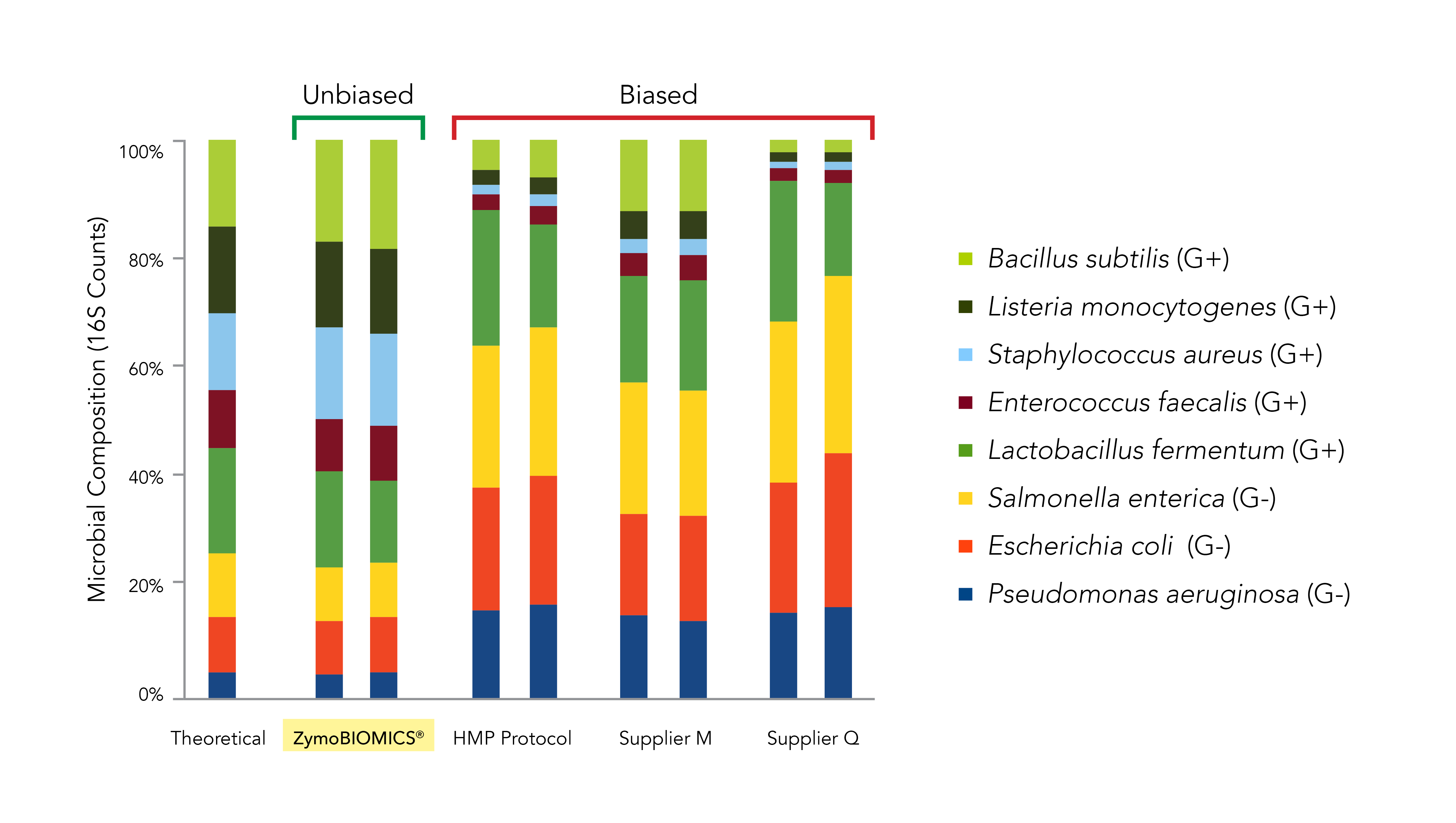
- Sinha R, Abnet CC, White O, Knight R, Huttenhower C: The microbiome quality control project: baseline study design and future directions. Genome Biol 2015, 16:276.
- Hsieh YH, Peterson CM, Raggio A, Keenan MJ, Martin RJ, Ravussin E, Marco ML: Impact of Different Fecal Processing Methods on Assessments of Bacterial Diversity in the Human Intestine. Frontiers in microbiology 2016, 7:1643. 13.
- Vishnivetskaya TA, Layton AC, Lau MC, Chauhan A, Cheng KR, Meyers AJ, Murphy JR, Rogers AW, Saarunya GS, Williams DE et al: Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS microbiology ecology 2014, 87(1):217-230. 14.
- Hart ML, Meyer A, Johnson PJ, Ericsson AC: Comparative Evaluation of DNA Extraction Methods from Feces of Multiple Host Species for Downstream Next-Generation Sequencing. PloS one 2015, 10(11):e0143334. 15.
- Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P, Thomson JM, Satsangi J, Flint HJ, Parkhill J et al: The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PloS one 2014, 9(2):e88982. 16.
- Sohrabi M, Nair RG, Samaranayake LP, Zhang L, Zulfiker AH, Ahmetagic A, Good D, Wei MQ: The yield and quality of cellular and bacterial DNA extracts from human oral rinse samples are variably affected by the cell lysis methodology. Journal of microbiological methods 2016, 122:64-72.
- Saey TH: Here is the poop on getting your gut microbiome analyzed. In: Science News. vol. 2017; 2014.
- Farkaš V, Takeo K, Maceková D, Ohkusu M, Yoshida S, Sipiczki M. Secondary cell wall formation in Cryptococcus neoformans as a rescue mechanism against acid-induced autolysis. FEMS Yeast Research, 2009, 9(2): 311-320
- Costea et al. Towards standards for human fecal sample processing in metagenomic studies. Nature Biotechnology(2017) 11:1069-1076


