Extracting DNA from plant leaves or seeds, while not a new concept, is the key to developing the plant-based technologies of the future. Plant DNA acts as the input for many downstream molecular biology techniques, including PCR and NGS, and nucleic acid extractions are foundational to the workflows of many academic researchers and commercial breeding programs. In modern agrobiotechnology and agrigenomics, generating accurate and actionable data for agricultural research, from finding pain management alternatives for opioids to global food security to mitigating the effects of climate change, relies on high-quality plant DNA, making the techniques, and the time and cost needed to execute them, important for many plant researchers.1-3
The Fundamental Steps for Plant DNA Extraction
In general, plant DNA extraction and purification can be divided into six steps: 1) tissue disruption/homogenization, 2) cell lysis in DNA extraction buffer, 3) separation of DNA from other cellular components, 4) DNA precipitation, 5) DNA washing, and 6) DNA collection/resuspension for downstream processing.
While it sounds biochemically simple, the nature of the plant starting material can present unique technical challenges that can be difficult to overcome for even experienced scientists. For that reason, several different plant DNA extraction methods have been developed and used for a variety of applications.
The Different Methods for Plant DNA Extraction
For decades, the conventional protocol for efficient, reliable DNA extraction from plants has been cetyltrimethyl ammonium bromide (CTAB) extraction, first developed in 1987.4,5 Over the years, technical modifications have been made to the protocol to make it more applicable to different plant species.6 In addition, protocols that incorporate a bead beating step have proven particularly useful for plant species with thick cell walls that are difficult to break apart.7
Column-based procedures using a silica membrane, under conditions that promote DNA binding, have also emerged. Similarly, magnetic beads are used for plant DNA extraction and (like column-based procedures) allow for efficient washing and purification of DNA.
Considerations for Choosing a Plant DNA Extraction Method
With a variety of DNA extraction methods and protocols to choose from, it can be difficult to know which is right for your experimental application and plant species. Here are some considerations for selecting a plant DNA extraction method.
Ease of Use and Length of Protocol
CTAB has been the conventional method of choice for plant DNA extraction, but it requires a large amount of equipment, reagents, and a significant number of labor-intensive steps.8 For that reason, it can be time consuming for a researcher to perform with a small number of samples and challenging to process a large number of samples. This method can take even skilled researchers multiple hours to perform.
Column- and/or magnetic bead-based protocols are much simpler to use and are commercially available as kits, with most reagents pre-prepared. In addition, the only equipment required is a centrifuge or magnetic stand and a bead beater (if working with plant tissue that is difficult to disrupt). These two methods can also be used for the implementation of scalable, automated workflows, which would not be possible with the many hands-on steps required for performing CTAB protocols. Some manual, column-based procedures can even be performed in as little as 15 minutes.
Lysis Efficiency
Lysis of plant cells is necessary for the extraction of DNA and separation from all other cellular components. Therefore, efficiency of cell (and nuclei) lysis is directly related to the quantity of DNA extracted. There can be significant variation in lysis efficiency across samples for those performing the CTAB protocol, as lignified and thick plant cell walls can be resistant to disruption by CTAB.4 Bead beating methods – which use physical force and agitation to disrupt plant cell walls – have helped to overcome these challenges, providing a more homogeneous and consistent tissue disruption technique. Bead beating has also enabled more efficient lysis for difficult-to-lyse plant species.
Removal of Downstream Inhibitors
A wide spectrum of polysaccharides, polyphenols, lipids, and other secondary metabolites can interfere with DNA extraction, contaminate purified DNA, and inhibit enzyme function, making downstream experiments challenging.9 In principle, CTAB protocols (when used in conjunction with additives such as polyvinylpyrrolidone) can remove many of these compounds, yet in practice, it can be extremely difficult to remove all contaminants. Many CTAB protocol modifications have been published, especially for plant species that are rich in these problematic compounds.9 However, Zymo Research's OneStep PCR Inhibitor Removal Kits are specially designed to remove common PCR inhibitors like polyphenolic compounds from even the most impure preparations, all in an efficient one-step procedure.
DNA Quality
Poor quality DNA can limit its use for downstream molecular analysis. For instance, using low-quality DNA for NGS can result in unreliable data or failed sequencing runs. High-quality DNA is typically characterized by the presence of high molecular weight fragments, which can be assessed by agarose gel electrophoresis or the use of a Bioanalyzer. In addition, it will generally be free of RNA, salts, phenol, and common secondary metabolites, which can be evaluated using a spectrophotometer and looking at 260 nm/280 nm and 260 nm/230 nm absorption ratios. As mentioned above, column- or magnetic bead-based protocols enable more efficient washing than protocols that use phase separation, DNA precipitation, and pelleting, as in the CTAB protocol.
DNA Loss
The downstream analysis might require a minimum amount of DNA, therefore, the quantity of DNA that will be extracted is also an important consideration. The CTAB protocol is prone to DNA loss during phase separation, DNA pelleting, and washing. While column-based procedures, although often lauded for the isolation of high-quality and inhibitor-free DNA, can also lead to significant reduction in overall yield due to the low binding capacity of the column and/or excessive washing.
Use of Hazardous Reagents
Phenol and chloroform are required for CTAB DNA extraction and are widely used for other nucleic acid extraction protocols. Yet working with them should be approached with caution.10,11 Both chemicals are listed by the Occupational Safety and Health Administration (OSHA) as hazardous to human health. Phenol can be absorbed through the skin and mucous membranes and can cause severe burns. Chloroform is a carcinogen and toxic if inhaled or swallowed. Prolonged exposure to phenol/chloroform should be avoided and proper safety precautions and disposal procedures should be followed according to OSHA and other regulatory bodies. Column- and magnetic bead-based procedures avoid the use of these chemicals, a major appeal for many researchers.
A solution for extracting DNA from Plant Leaves & Seeds
Zymo Research’s Quick-DNA Plant/Seed Kits completely trivializes the biggest challenges with traditional CTAB extraction and other DNA extraction methods. There is no need for hazardous phenol or chloroform and many problematic secondary metabolites are removed in a single centrifugation step.
In a nutshell, the plant tissue is lysed by efficient mechanical disruption through bead beating, followed by a series of centrifugation steps using spin-columns to purify the DNA and remove downstream inhibitors (Figure 1). The resulting eluted DNA is ideal for many downstream molecular biology and sequencing applications. This simple process can be done in as little as 15 minutes – in conjunction with tissue grinding and bead bashing – on a variety of plant samples, including leaves, stems, buds, flowers, fruit, and seeds, giving you a straightforward path to high-quality DNA.
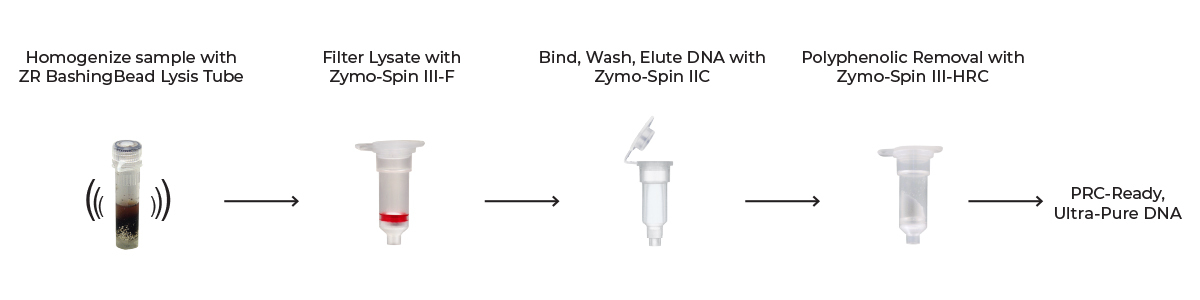
The kit has been validated with crops that have been traditionally difficult to extract high-quality DNA from.12-15 This includes plants like Theobroma cacao, which contains high levels of anthocyanins and other polyphenolics that interfere with some nucleic acid purification protocols and many downstream reactions like PCR. For that reason, Mars, Inc. (and other agricultural pioneers) has incorporated it’s use into several of its core agrigenomics workflows.
The Story of Mars, Inc. and Plant Extraction
A 2017 study, in collaboration with an international group of researchers, used the Quick-DNA Plant/Seed 96 Kit to implement a high-throughput isolation protocol for extracting DNA from a total of 3,733 individual Theobroma cacao plants (representatives of a panel of 148 cacao clones), selected for improved productivity and resistance to black and frost pod disease. They performed SNP genotyping across 90 different markers with the purified DNA.13 The efficiently extracted high-quality DNA enabled the team to accurately perform genome wide association studies (GWAS) that identified six markers significantly associated with disease resistance and driving continued improvement of cacao productivity.
Later on, in a 2018 study, the Mars, Inc. team, along with another international team, used the Quick-DNA Plant/Seed kits to extract genomic DNA and sequence 200 T. cacao genomes to reconstruct the path of the plants domestication, which occurred approximately 3600 years ago.14 Taken together, our plant and seed DNA extraction kit has helped further the understanding of a crop that continues to play a central role in modern agriculture and food production.
The future of climate change solutions, sustainable global food security, and more begins with high-quality DNA extraction, and Zymo Research has the tools to help your lab deliver the next big agricultural breakthrough.
Citations
- Todd DA, Kellogg JJ, Wallace ED, et al. Chemical composition and biological effects of kratom (Mitragyna speciosa): In vitro studies with implications for efficacy and drug interactions. Sci Rep. 2020;10(1). doi:10.1038/S41598-020-76119-W
- Sedeek KEM, Mahas A, Mahfouz M. Plant Genome Engineering for Targeted Improvement of Crop Traits. Front Plant Sci. 2019;0:114. doi:10.3389/FPLS.2019.00114
- Varshney RK, Bansal KC, Aggarwal PK, Datta SK, Craufurd PQ. Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 2011;16(7):363-371. doi:10.1016/j.tplants.2011.03.004
- Abdel-Latif A, Osman G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods. 2017;13(1):1-9. doi:10.1186/S13007-016-0152-4
- Doyle J, Doyle J. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem Bull. 1987;19(1):11-15.
- Aboul-Maaty NAF, Oraby HAS. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull Natl Res Cent. 2019;43(25). doi.org/10.1186/s42269-019-0066-1
- Angeles JGC, Laurena AC, Tecson-Mendoza EM. Extraction of genomic DNA from the lipid-, polysaccharide-, and polyphenol-rich coconut (Cocos nucifera L.). Plant Mol Biol Report. 2012;23(3):297-298. doi:10.1007/BF02772760
- CTAB PROTOCOL FOR ISOLATING DNA FROM PLANT TISSUES. Zymo Research: https://www.zymoresearch.com/blogs/blog/ctab-protocol-for-isolating-dna-from-plant-tissues
- Sahu SK, Thangaraj M, Kathiresan K. DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol. ISRN Mol Biol. 2012;2012:205049. doi:10.5402/2012/205049
- 22. Safe Use of Phenol | Safety Services. UC - Davis Safety Services website: https://safetyservices.ucdavis.edu/safetynet/safe-use-of-phenol. Published March 26th, 2020. Accessed October 27th, 2021.
- Report on Carcinogens, Fourteenth Edition. National Toxicology Program website: https://ntp.niehs.nih.gov/ntp/roc/content/profiles/chloroform.pdf. Published November 3rd, 2016. Accessed October 27, 2021.
- Kamber T, Malpica-López N, Messmer MM, et al. A qPCR Assay for the Fast Detection and Quantification of Colletotrichum lupini. Plants. 2021;10(8):1548. doi:10.3390/PLANTS10081548
- Romero Navarro JA, Phillips-Mora W, Arciniegas-Leal A, et al. Application of Genome Wide Association and Genomic Prediction for Improvement of Cacao Productivity and Resistance to Black and Frosty Pod Diseases. Front Plant Sci. 2017;8:1905. doi:10.3389/fpls.2017.01905
- Cornejo OE, Yee MC, Dominguez V, et al. Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process. Commun Biol. 2018;1:167. doi:10.1038/s42003-018-0168-6
- Garfinkel AR, Otten M, Crawford S. SNP in Potentially Defunct Tetrahydrocannabinolic Acid Synthase Is a Marker for Cannabigerolic Acid Dominance in Cannabis sativa L. Genes. 2021;12(2):228. doi:10.3390/GENES12020228
