
High Quality ATAC-Seq Libraries: Zymo-Seq ATAC Library Kit
from as little as 50,000 cells
Discover, Explore, and Map the Epigenetic Landscape with the Zymo‑Seq ATAC Library Kit
Low Input: Assay requires only 50,000 cells to prepare libraries.
Sample Flexibility: Quality data produced from both fresh and frozen sample types.
Streamlined Workflow: Minimal hands-on time increases assay speed while reducing potential for error
Popular Applications of ATAC-Seq

Cancer Research
Developmental Biology

Biomarker Discovery
Disease Studies

Drug Discovery
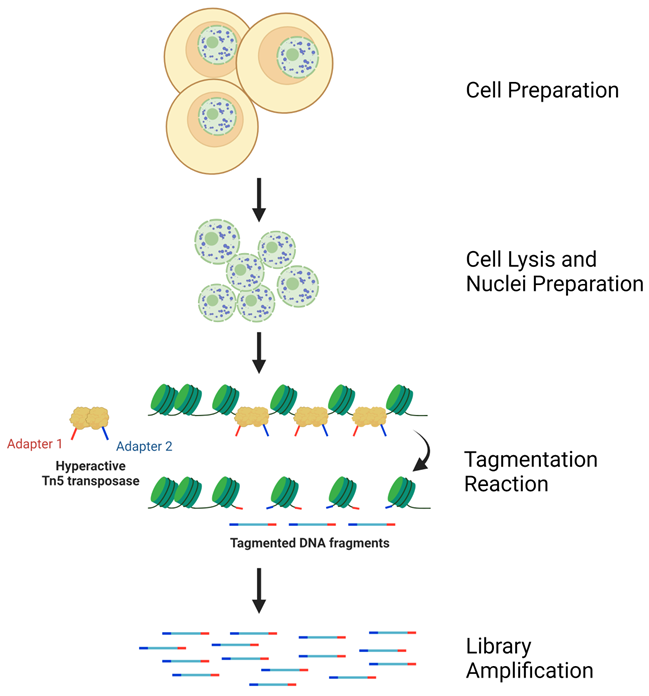
Lighting-Fast Solution with Ready to Use Buffers
Customer Testimonial
Powerful Tool to Study Open Chromatin with Outstanding Consistency
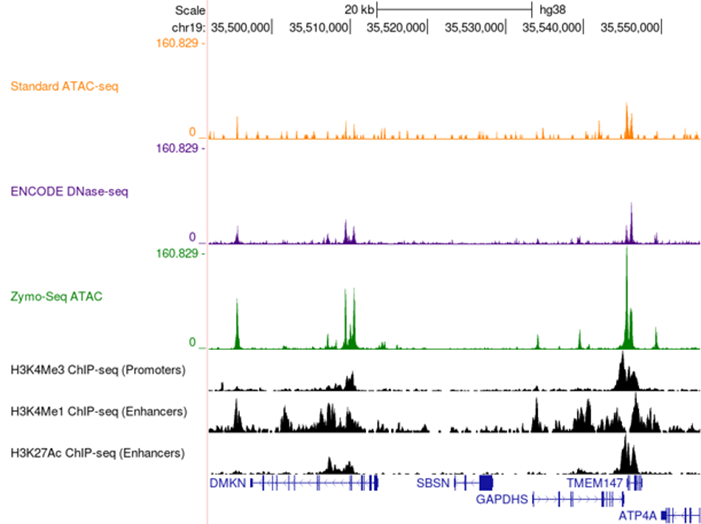
Browser tracks depicting GM12878 ATAC-Seq assay using the Zymo-Seq™ ATAC Library Kit. Peaks overlap at the same sites identified by DNase-Seq in the ENCODE project as well as the standard ATAC-Seq protocol showing both quality and consistency.

The Zymo-Seq™ ATAC Library Kit delivers top tier replicate correlation with Pearson’s R consistently greater than 0.95. This high correlation depicts the reproducibility and consistency of the assay.
