Are Liquid Biopsies the Future of Diagnostics?
In the constantly improving field of clinical diagnostics, a revolution is occurring in the form of liquid biopsies and cell-free nucleic acid (cfNA) analysis. Liquid biopsies are a non/less-invasive method to analyze health status from biological fluid samples (serum, saliva, urine, and other fluid sample types). Typically, they require small amounts of starting material (<10 mL), are minimally-invasive and simple to perform. Compared to liquid biopsies, surgical biopsies can be very invasive, time consuming, painful, and are limited in scope.
The significance of liquid biopsies lies in their ability to detect early disease with just a simple blood draw. As technologies improve the detection of specific disease biomarkers, earlier adoption of more effective targeted treatments is possible. This potential has been gaining a lot of attention, leading to significant investments by companies like Illumina and Roche.
Cell-free Nucleic Acids
It has long been recognized that biological fluids contain DNA and RNA that is not contained in cells. It is now known that these cell-free nucleic acids are released into biological fluids from apoptotic and necrotic cells or secreted from healthy and diseased cells via microvesicles such as exosomes [1]. The presence of cfNA in biological liquids has been long recognized, but its function is poorly understood, as practical uses have been hampered by insufficient isolation and analytical methods [2]. For example, cfNAs have been linked to cancer since their discovery in 1948 [3], but due to technical limitations, it took almost 20 years for a link between cell-free DNA (cfDNA) and cancer to be established and widely supported.
It took another 20 years to define tumor-derived cfDNA within samples taken from cancer patients [4]. However, with the inception of the Human Genome Project and recent technological advances in genome analysis, the field of cell-free nucleic acid research has grown rapidly and cell-free RNA (cfRNA) analysis is starting to gain recognition as a source of important biomarkers [5].
The use of cell-free nucleic acid detection has expanded beyond cancer detection. Liquid biopsy methods are now being evaluated for diagnostic effectiveness in disease and pre-natal genetic testing. If proven effective, these techniques will allow for safer and earlier in vivo detection of various inherited genetic diseases. The goal is to implement personalized medical interventions in early stages of disease for a better chance of positive effects on health outcomes. Many tests already widely used with cell-free fetal DNA (cffDNA) include a first trimester Down syndrome screen, amniotic fluid analysis, and chromosomal karyotype analysis.
Many applications are currently being evaluated as a less-invasive method to monitor patient health. These include testing for fetal aneuploidy, monitoring the status of organ transplants, blunt trauma and burn recovery progression, and detection of sepsis and septic shock in patients [6]. In addition to these, research is underway to use cfNA for early diagnosis of genetic disorders such as β-thalassemia and sickle cell anemia [6]. Earlier detection will lead to earlier treatment of many of these disorders, which will improve treatment outcomes and prevent comorbidities from developing.
Sample Diversity and Purification Issues
Numerous biofluids such as blood, cerebrospinal fluid (CSF), saliva, amniotic fluid, and urine [4, 6-9] contain readily detectable cfNA populations. However, each of these sample sources produce specific challenges during collection, handling, and extraction that must be addressed to obtain accurate profiles. Cell-free DNA is most commonly isolated from the serum or plasma fraction of blood, however extraction methods must also be able to address a wide range of sample types.
The most readily available methods to collect cfDNA have been restricted to commercially available extraction kits or homemade methods. These methods may not be optimized and often use outdated technologies. Cell-free biofluids are challenging to work with due to their high protein content and low amount of cfNA in relatively large amounts of liquid. This is why traditional cfNA extraction technologies have been so limited in isolation efficiency, sample compatibility, and have been unable to isolate both cfDNA and cfRNA from the same input sample.
To address this issue, Zymo Research produced a revolutionary kit designed to easily process samples from liquid biopsies, all in one quick, simple workflow. The Quick-cfDNA/cfRNA™ Serum & Plasma Kit contains protocols that make it easy to isolate cfDNA from even the most difficult sample types (Figure 1).
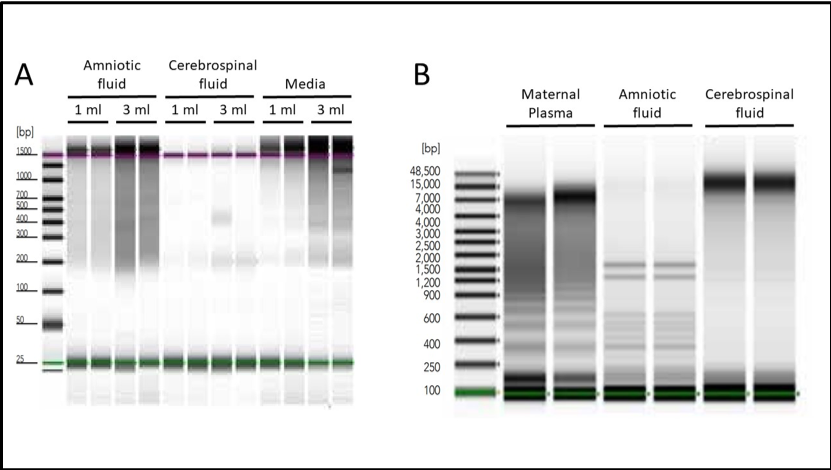
Detection and Quantification of Cell-Free Nucleic Acids
Liquid biopsies often yield relatively miniscule amounts of total cell-free nucleic acid content. For example, plasma samples typically contain only 1-100 ng/mL of cfRNA, whereas whole blood contains ~2-10 µg/mL total RNA. Alternatively, plasma samples typically contain 1-10 ng/mL cfDNA whereas whole blood contains 30-70 µg/mL total DNA.
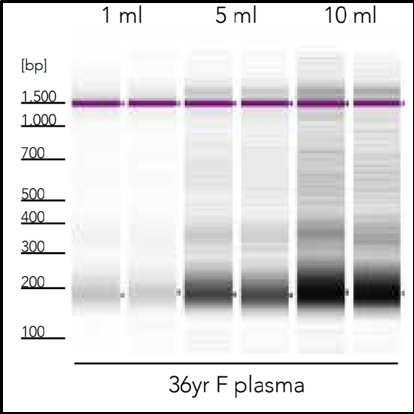
Because of minimal amounts of total cell-free nucleic acid content, it is often necessary to process large volumes of the cell-free fraction of a biofluid sample to recover a quantifiable and workable cfNA (Figure 2). The low molecular mass of cfNA also produces its own unique challenges in the isolation process, especially for microRNA which has average length of 22 nt, since most silica-based purification technologies are optimized to bind nucleic acid of a much higher molecular weight. These challenges are what inspired Zymo Research to create the Quick-cfDNA/cfRNA Serum & Plasma Kit, which allows fast, consistent recovery of the highest amount of cfNA of any technology available. (Figure 3).
Cell-free Nucleic Acid Profile Analysis
Another challenge facing cfNA analysis is the robustness of samples when initiated into sequencing pipelines such as RNA sequencing (RNA-Seq). Along with the relatively low amount of nucleic acid, microRNAs are often lost during purification which leads to profile bias. This is can be due to the use of non-optimal extraction methods that produce biased cfNA recovery. For example, three commercial methods of cfRNA extraction from plasma demonstrates difference in the cell-free microRNA yield for hsa-miR-16-5p in the figure below (Figure 3).
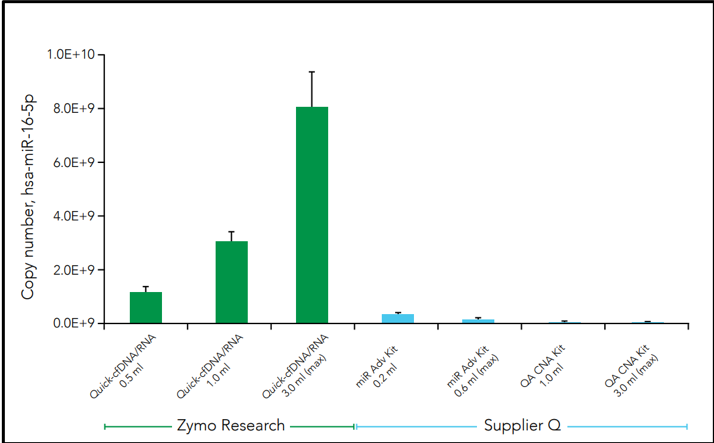
Further comparative analysis of commercial kit cfRNA extracts followed by RNA-seq analysis demonstrates that the Quick-cfRNA Serum & Plasma kit yielded higher quality reads, better alignment to sequence databases, and achieved recovery of 463 additional microRNA species not recovered by the supplier Q kit (Table 1).

Conclusion
The field of liquid biopsy is rapidly growing due to the massive technological revolution in genomics over the past 30 years. The discovery of cell-free nucleic acids has allowed for doctors to monitor cancer and other disease states in human adults as well as detect complications early on in pregnancy. The advent of analytical methods for cell-free nucleic acid detection has allowed for significantly safer disease monitoring and pregnancy testing. This is because rather than a surgical biopsy or amniocentesis physicians can get all the information from a simple blood draw. With these tools in place, scientists and doctors can work together to bring us to a healthier and safer future through early diagnosis and rapid disease monitoring.
References:
[1] Dominguez-Vigil IG, Moreno-Martinez AK, Wang JY, Roehrl MHA, Barrera-Saldana HA. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2018;9:2912-22.
[2] Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24.
[3] Mandel P, Metais, P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil. 1948;142:241-3.
[4] Bryzgunova OE, Laktionov PP. Extracellular Nucleic Acids in Urine: Sources, Structure, Diagnostic Potential. Acta Naturae. 2015;7:48-54.
[5] Zaporozhchenko IA, Ponomaryova AA, Rykova EY, Laktionov PP. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn. 2018;18:133-45.
[6] Soltani M, Nemati M, Maralani M, Estiar MA, Andalib S, Fardiazar Z, et al. Cell-free fetal DNA in amniotic fluid supernatant for prenatal diagnosis. Cell Mol Biol (Noisy-le-grand). 2016;62:14-7.
[7] Connolly ID, Li Y, Pan W, Johnson E, You L, Vogel H, et al. A pilot study on the use of cerebrospinal fluid cell-free DNA in intramedullary spinal ependymoma. J Neurooncol. 2017;135:29-36.
[8] Hui L, Bianchi DW. Cell-free fetal nucleic acids in amniotic fluid. Hum Reprod Update. 2011;17:362-71.
[9] Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17:3.
[10] Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics. 2014;15:29.


