Tips & Tricks For RNA Isolation
RNA extraction is a notoriously difficult endeavor. Common challenges include RNA degradation, low yield and/or purity, and DNA contamination. Additionally, different sample types have their own unique features that require special attention. For example, microbes (e.g. gram positive/negative bacteria, fungi, archaea, etc.) can have tough cell walls that are refractory to chemical and enzymatic lysis. Other sample types such as feces and plants contain inhibitors (e.g. polyphenolics, humic/fulvic acids, tannins, etc.) that can co-precipitate with RNA and can inhibit downstream analysis such as RT-qPCR.
Below, top scientists have shared their best RNA isolation tips, specifically best practices for maximizing the recovery of high quality, DNA-free RNA.
Stabilizing RNA After Collection
RNA can be unstable and highly susceptible to degradation. Many samples contain high levels of RNases which rapidly and efficiently degrade RNA. To minimize this, it is best to stabilize samples at the moment of collection. Common methods for sample stabilization include snap freezing with liquid nitrogen, dry-ice ethanol baths, or storage in a -80°C freezer. However, these approaches have drawbacks, such as freeze-thaw damage of nucleic acids, and not all researchers have immediate access to these methods at the time of sample collection.
Best RNA Stabilization Methods:
- Immediate solubilization in a lysis buffer that inactivates RNases (e.g. TRIzol®, RNA Lysis Buffer, etc.). Samples can then be processed immediately or stored frozen.
- Submersion in a stabilization reagent (e.g. DNA/RNA Shield) that inactivates nucleases and protects nucleic acids at ambient temperature for extended periods of time. This is particularly helpful for researchers that are collecting samples in the field or working with precious patient samples (e.g. tissue biopsies, whole blood, etc.).
Ensure Complete Sample Lysis
During RNA extraction, the best way to maximize RNA yield and quality is to ensure complete sample lysis. However, not all samples will be susceptible to the same lysis regimen. For example, blood cells (e.g. lymphocytes, PBMCs, etc.) and microbial cells tend to be more difficult to efficiently lyse. Simply adding a detergent-based lysis buffer may not always be sufficient. To help improve lysis, it is helpful to pair the lysis buffer with a mechanical lysis step (e.g. bead beating) or include an enzymatic lysis step upstream (e.g. proteinase K, lysozyme, etc.) (Figure 1).
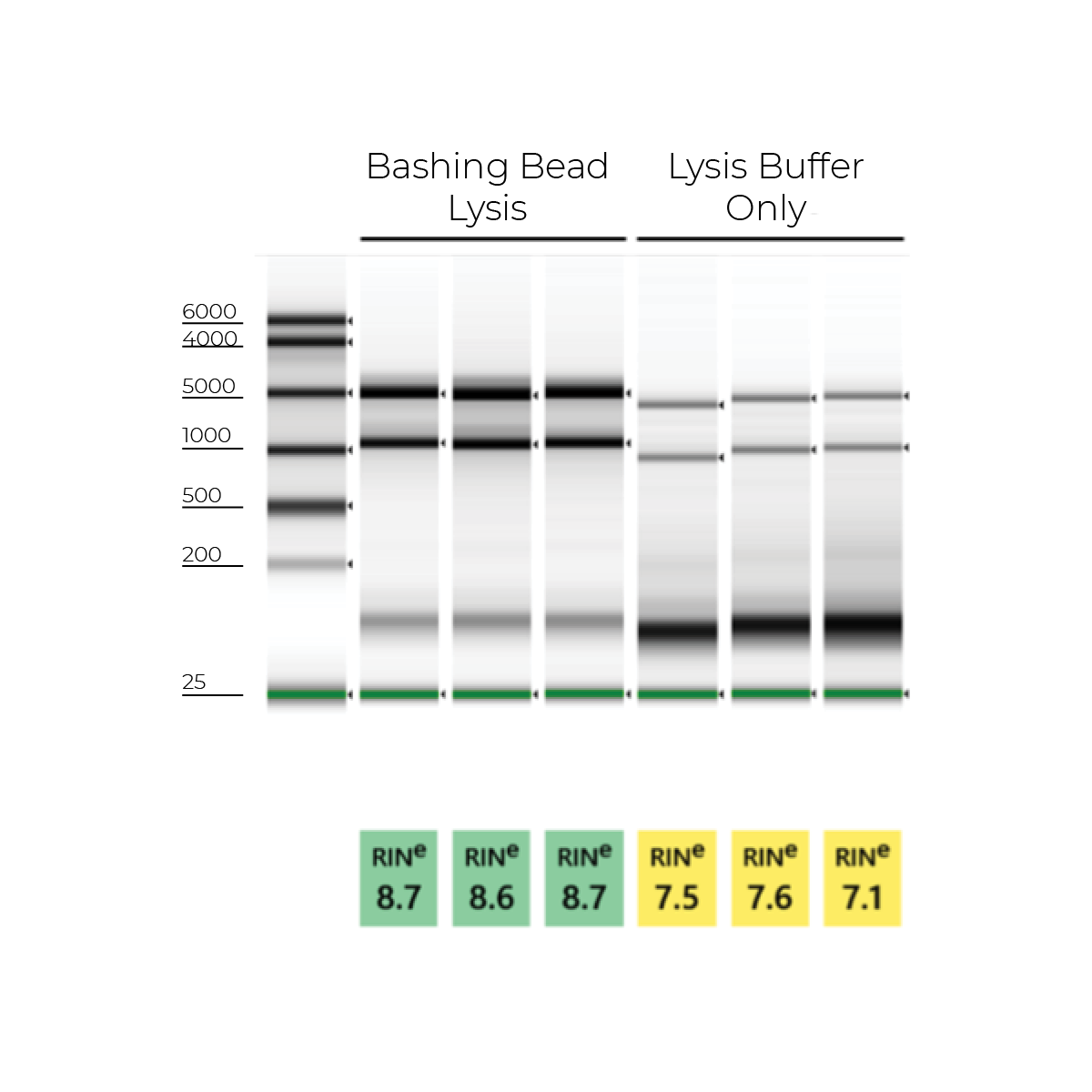
Furthermore, it is important to note that complete sample lysis also helps the extraction procedure run smoothly. Incomplete lysis with column-based extraction methods can cause column clogging, buffer carryover, DNA contamination, and incomplete elution. With all of this in mind, Zymo Research has gone to great lengths to optimize the most efficient lysis regimens for all common sample types.
How to Eliminate DNA Contamination
Another common problem associated with RNA extraction is DNA contamination. The presence of DNA can skew UV/VIS based quantification methods (e.g. Nanodrop), artificially increasing the RNA quantification higher than what it really is. Plus, it can also result in false readings in more sensitive downstream applications (e.g. RNA-seq). To ensure that you are getting the most accurate quantification measurements and to avoid any discrepancies in downstream analysis, it is important to eliminate any DNA carryover. This can be done in a variety of ways (e.g. TRIzol® phase separation, DNA removal columns, and DNase treatment).
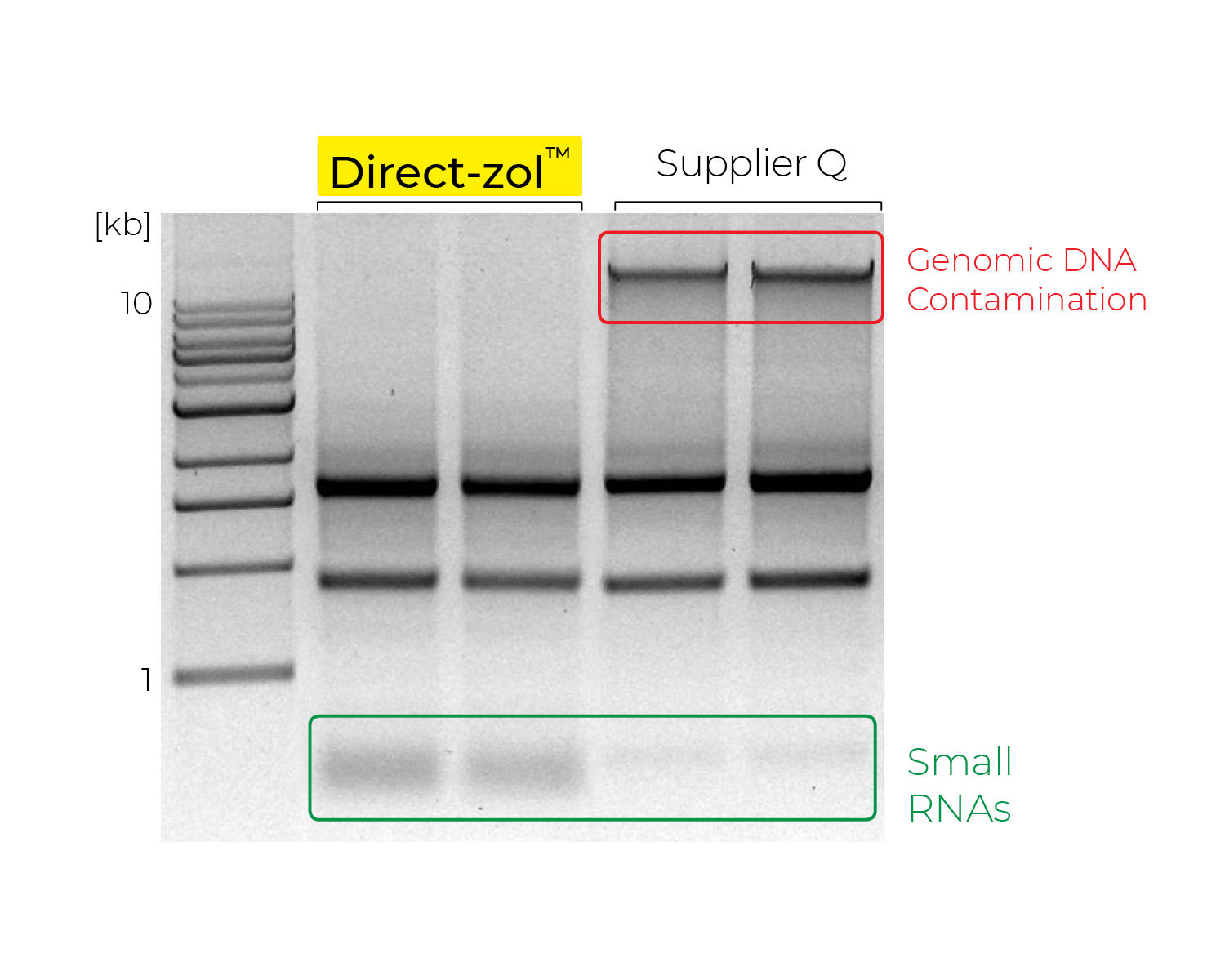
The fastest method for confirming the presence of DNA is to visualize the RNA samples (e.g. agarose gel, Agilent TapeStation®, etc.) and look for any high molecular weight fragments above the 28S ribosomal RNA band (Figure 2). Other methods, such as qPCR or Qubit, can also be used and are preferable if you are performing downstream applications with sensitivity to DNA contamination.
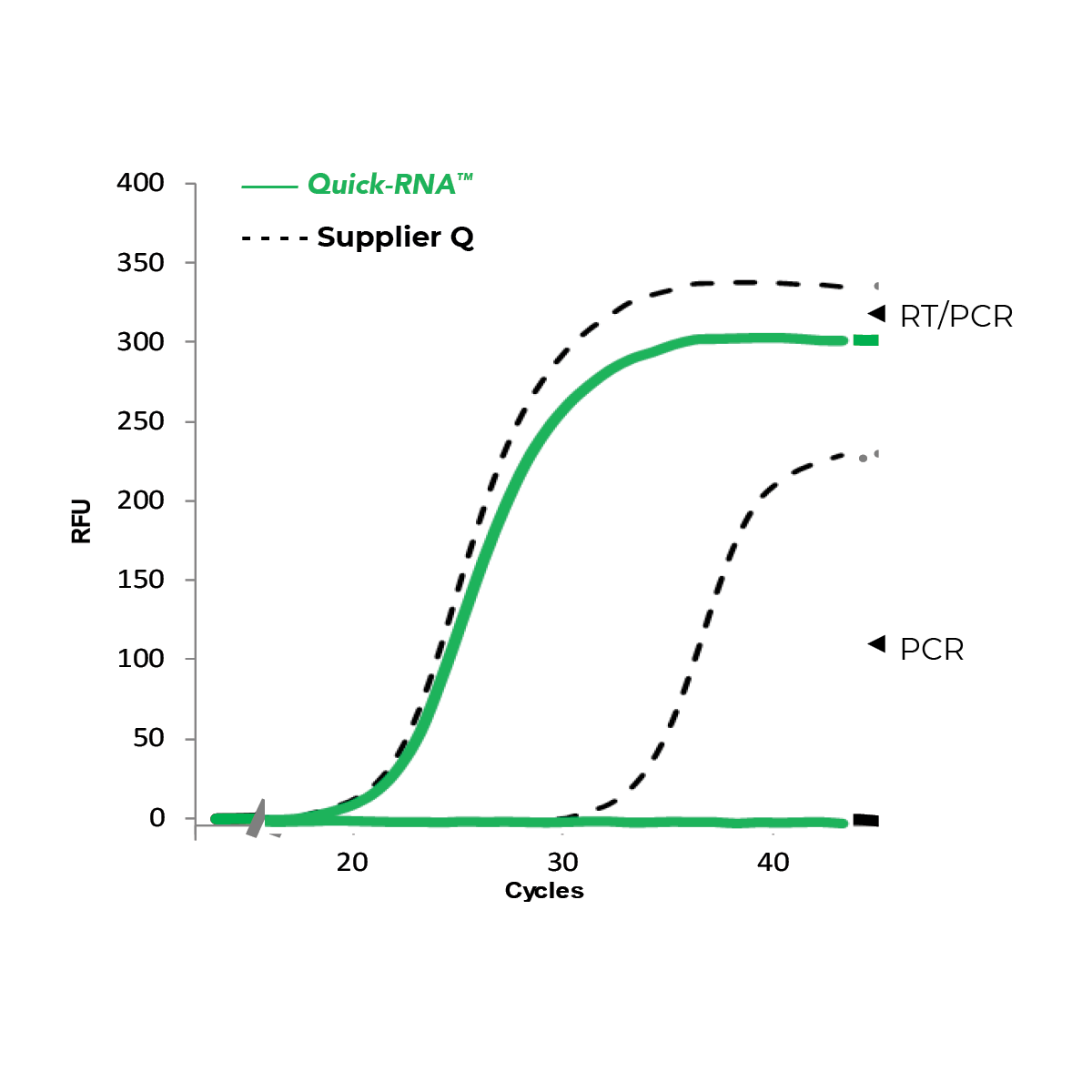
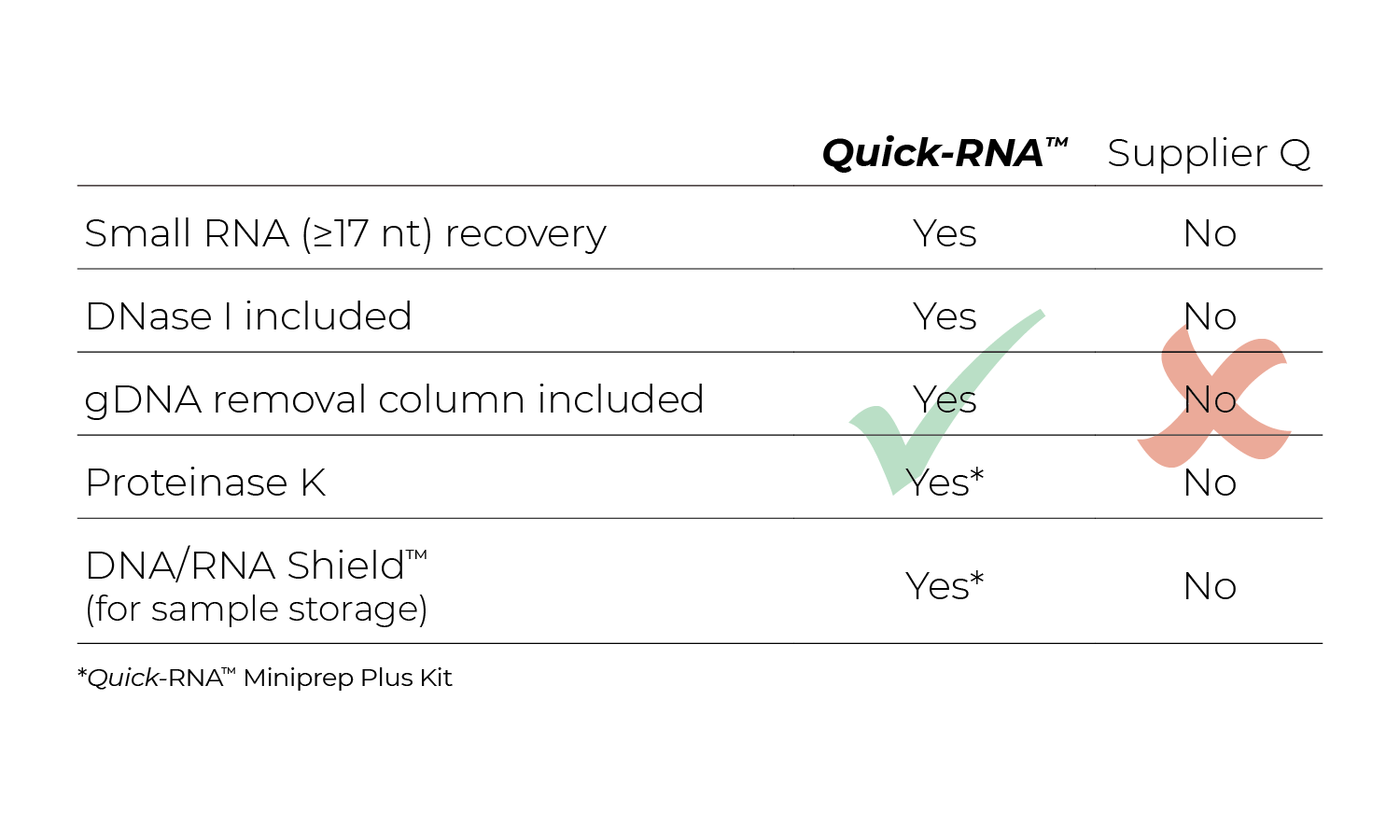
Zymo Research has developed its RNA extraction kits with novel buffer and columns systems that bind and extract RNA while eliminating contaminating DNA. In comparison, many other products on the market are co-purification based, and thus retain high levels of DNA contamination. As an added value, the best RNA isolation kits include a DNase I set for on-column treatment. This removes the need for a post-extraction DNase treatment and clean-up steps as well as streamlines the process from extraction to downstream application. Figure 3 below illustrates how effective the Zymo on-column DNase treatment is based on the lack of DNA amplification via PCR.
Choosing the Best RNA Isolation Kits for the Job
Knowing that RNA isolation is not a one-size fits all process, Zymo Research has developed a wide array of the best RNA isolation kits that are tailored to fit most sample types. Plus, most of these RNA isolation kits are available in various formats (i.e. Microprep, Miniprep, 96-well, Magnetic Bead, etc.) to suit different input amounts and throughput.
As a reference, Table 1 shows specific sample types with the recommended RNA extraction method/kit from Zymo Research. Also, the Zymo Research technical support team is always available to help ensure successful RNA extractions in any workflow.
| Sample Type | Recommended Kit |
|---|---|
| Samples in TRIzol® | Direct-zol RNA Kits |
| Cells, Tissue, Blod and other Biological Fluids | Quick-RNA Kits |
| Viral RNA | Quick-RNA Viral Kit |
| Whole Blood | Quick-RNA Whole Blood |
| Urine | ZR Urine RNA Isolation Kit |
| Tissue Sections | Pinpoint Slide RNA Isolation System |
| FFPE Samples | Quick-RNA FFPE Miniprep |
| cfRNA from Serum, Plasma, CSF, and Amniotic Fluid | Quick-cfRNA Serum & Plasma Kit |
| Feces, Soil, Biofilm, Water, and Culture | ZymoBIOMICS RNA Miniprep Kit |
| Plant Samples | Quick-RNA Plant |
| Microbial Cultures | Quick-RNA Fungal/Bacterial Kits |
| Insects (mosquitos, bees, drosophila, ticks, etc.) | Quick-RNA Tissue/Insect Kit |
| RNA from Enzymatic Reactions, aqueous phase, etc. | RNA Clean & Concentrator Kits (RCC) |
| RNA from Agarose Gels | Zymoclean Gel RNA Recovery Kit |
| RNA containing PCR inhibitors (polyphenolics, humic/fulvic acids, tannins, etc.) | OneStep PCR Inhibitor Removal Kit |
| Sample Collection and RNA Stabilization | DNA/RNA Shield |
Mastering RNA Extractions
High-quality total RNA can be recovered from any sample type by keeping in mind three simple tips: ensure sample stabilization post collection, lyse the sample thoroughly and completely, and eliminate any potential DNA contamination. These tips will make it easier to recover RNA that is suitable for any downstream application and obtain downstream results that are accurate and reliable every time. Also, all-inclusive products, like those listed above, can simplify the extraction procedure and are supported by free technical support teams that can provide any additional recommendations and help anyone become an RNA extraction master!


